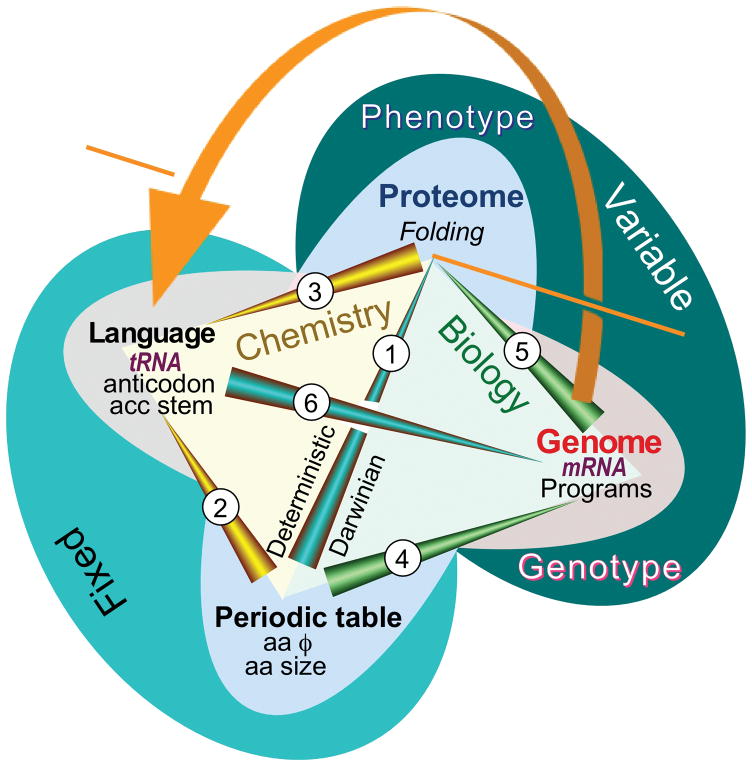
Figure 2.
Aminoacyl-tRNA synthetases and their cognate tRNAs furnish the reflexive elements necessary to translate the genetic code (orange arrow). The code connects their gene sequences, via their folded structures, to the enzymes that can enforce the coding rules in the codon table. Network analysis of the Central Dogma of Molecular Biology accommodates new evidence relating both tRNA identity elements and protein folding directly to the free energies of phase transfer equilibria of amino acid side chains [40,41,55]. Two of the nodes of a tetrahedron—physical properties of amino acids and the codon assignment table—reside in the realm of chemistry. They are “fixed” because they obey chemical equilibria. The other two nodes—gene sequences and protein folding—are dynamic processes that transcend chemistry because they are variable and form the basis for the evolution of diversity through self-organization and natural selection. The network connects RNA and Protein worlds via six edges: 1. Protein folding depends on both amino acid polarity and size [55], properties that play a role analogous to those of elements in chemistry, in that the amino acids form a kind of “periodic table” on which the folding of proteins is based. 2. tRNA bases encode amino acid size and polarity separately [41]. The acceptor stem codes for amino acid sizes; the bases of the anticodon code for amino acid polarities. Amino acid properties therefore dictate how tRNA bases are recognized by aminoacyl-tRNA synthetases. 3. tRNA codes are related to protein folding. Together, statements (1) and (2) imply that for the aminoacyl-tRNA synthetases, the code (and mRNA sequences) must define folded structures that bind specifically to particular tRNAs in order to read the language of genetics. The bi-directionality of this arrow illustrates a somewhat deeper self-referential element than those identified in molecular biology by Hofstadter as generators of complexity according to Gödel’s incompleteness theorem [179]. 4. Gene sequences (mRNA) are analogous to computer programs. Genetic instructions assemble amino acids according to their physical properties in ways that, when translated according to the programming language in tRNA (in 5), yield functional proteins (enzymes, motors, switches, regulators). 5. mRNA sequences (i.e., the genotype) determine amino acid sequences in proteins, and hence how amino acid sizes and polarities are exploited to produce different folded proteins. The spontaneous folding of amino acid sequences gives rise to functions (i.e., the phenotype) that ultimately determine whether or not a particular sequence survives natural selection. Changes accumulated in gene sequences result from selection acting on the phenotype. (4) and (5) localize how selection incorporates information about amino acid behavior into gene sequences, hence depict the evolutionary dimension in biology. 6. The evolution of mRNA sequences only makes sense in the context of the translation table (or programming language) established by the genetic code. (Adapted, with permission, from Carter, CW, Jr & Wolfenden, R. (2016) RNA Biology 13:145–151.)