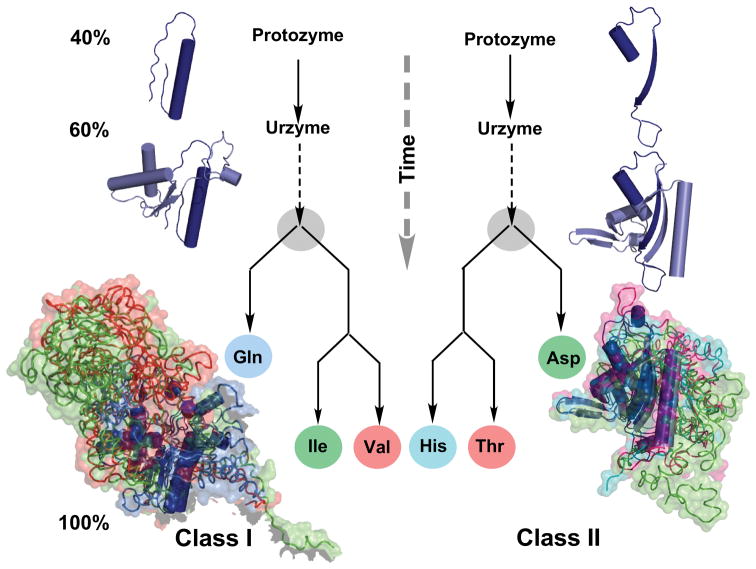
Figure 3.
Inferring protein family trees from molecular anatomies. Structures of three class I and class II aaRS have been rotated into a common orientation using their atomic coordinates and colored differently. α-Helical secondary structures are drawn as cylinders; extended β-structures as ribbons with arrows indicating their direction. Larger, more differentiated structures are drawn as noodles, surrounded by their surfaces. Differences in the recent structures at the bottom are highlighted by modules of one color that are absent in other structures. Such differences can be quantified and used to construct the genealogies in the center. Modules that are most similar in all three are colored dark blue and are inferred to be present in the common ancestor. Circles represent essentially modern aaRS. The three structures in each aaRS class are labeled with their three-letter abbreviations. There is consensus that they were present together with twelve other aaRS in the last universal common ancestor (LUCA) of all living organisms [180,181]. Novel results described here are the construction, expression, and experimental testing of ancestral forms called urzymes and protozymes, which are found, essentially without variation in all contemporary species and which retain substantial fractions—60% and 40% respectively—of the catalytic activity of the contemporary enzymes. The similarity between the class I and II genealogies is evidence that the two families evolved coordinately. (Courtesy of Carter Natural History CW, Jr. (2016) Natural History 125:28–33.)