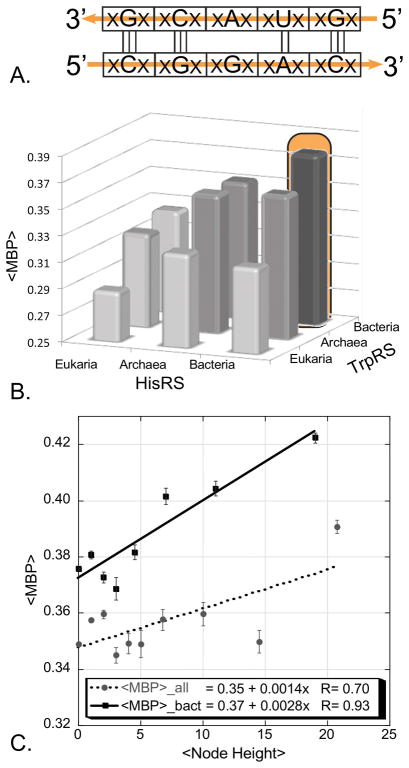
Figure 8.
Codon middle-base pairing (<MBP>) furnishes a new phylogenetic distance metric. A. Transient bi-directional coding is rapidly lost after the constraint is released. First and third bases lose complementarity much faster than the middle bases, which retain residual base-pairing into contemporary sequences. B. Comparison of middle codon-base pairing in all-by-all alignments of “Urgene” sequences excerpted from ~200 contemporary HisRS and TrpRS sequences [7]. C. Node-dependence of middle codon-base pairing in antiparallel alignments of middle bases from ancestral sequences reconstructed independently for the TrpRS and HisRS Urzyme multiple sequence alignments. Solid line is for bacterial sequences, dashed line is for all bacterial, archaeal, and eukaryotic sequences. (B and C adapted with permission from the Society for Molecular Biology and Evolution. Chandrasekaran, SN et. al. (2013) Mol. Biol. Evol. 30: 1588–1604.)