Figure 1.
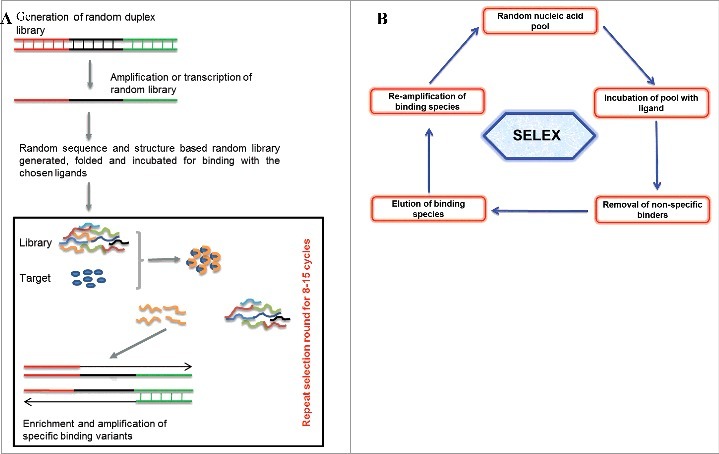
(1A) Representation of aptamer selection. Briefly random oligonucleotide libraries with diverse structural complexities are generated and incubated for ligand binding. Non-specific binders are removed and high affinity binders are amplified. The selection cycle are usually repeated around 5-20 cycles. The sequences and structural characterization from chosen binders are done to achieve the best binding aptamer. (1B) Basic strategy of SELEX protocol showing incubation of random library with chosen ligands, their incubation for binding, removal of non-specific binder and finally the enrichment of the best binders for chosen targets.
