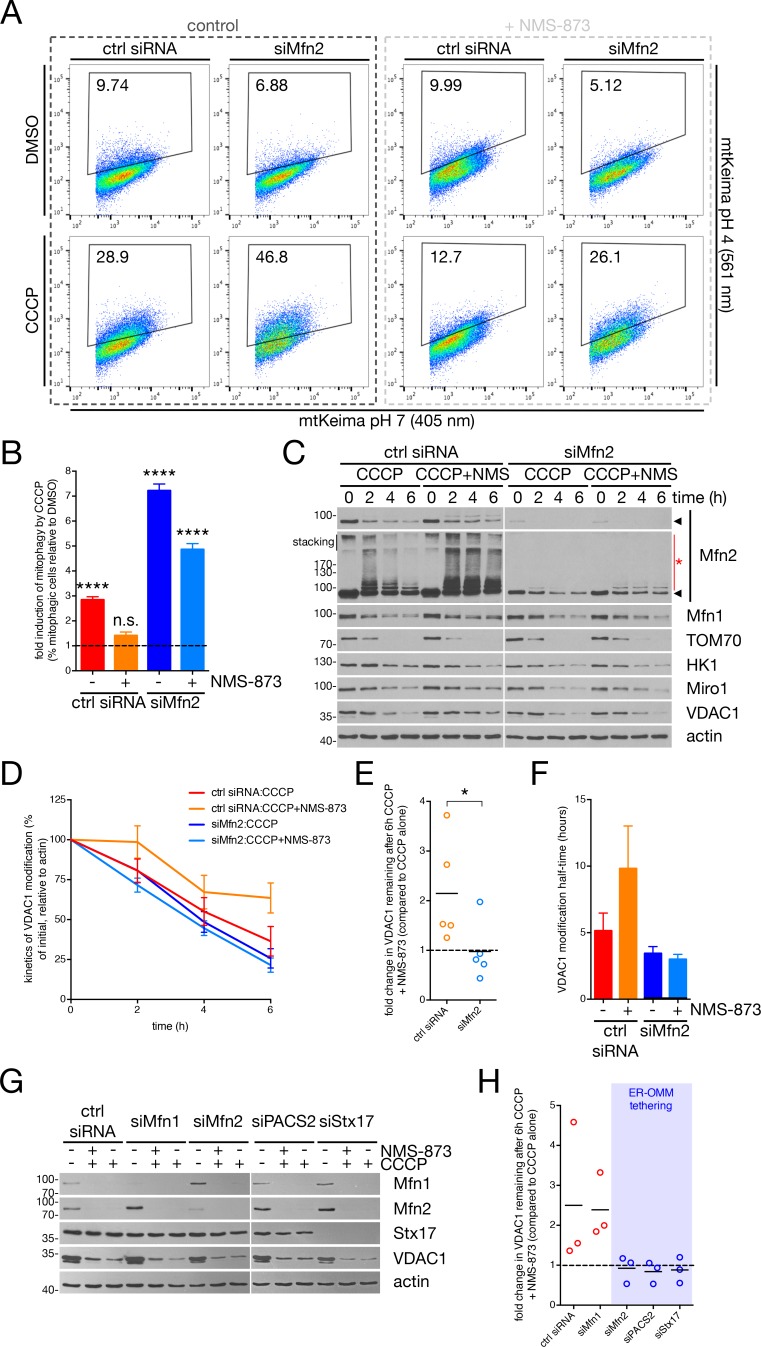
Figure 7. p97 and Mfn2 effect mitophagy through parkin substrate availability.
(A) U2OS:mtKeima cells were transfected with the indicated siRNA and GFP-parkin WT, and were treated with 20 μM CCCP (or DMSO) for five hours in the presence (dark grey box) or absence (light grey box) of 25 μM NMS-873. mtKeima fluorescence in GFP-positive cells was measured using flow cytometry by excitation at 405 nm (neutral pH) and 561 nm (acidified). The data are represented as scatter plots of fluorescence emission from excitation at both wavelengths. The gated area encloses cells undergoing mitophagy and the percentage of cells within this gate is indicated in the top-left corner of each plot. (B) Quantification of the percent of cells undergoing mitophagy in cells from (A), expressed as a ratio of CCCP-treated cells to those treated with DMSO. Bars represent mean ± SEM, n = 2 experiments. n.s., not significant; ****, p<0.0001. (C) Immunoblot analysis of U2OS:GFP-parkin cells, transfected with siRNA targeting Mfn2 (siMfn2) or control (ctrl siRNA), treated with 20 μM CCCP in the presence or absence of 25 μM NMS-873 over a period of six hours. (D) Immunoblot quantification of VDAC1 levels (relative to actin) from cells from (C). Bars represent mean ± SEM, n = 5 experiments. (E) The 6 hr time-point data from (D) is represented as a fold change in VDAC1 remaining when NMS-873 is added. Data points are represented on the graph, n = 5 experiments. *, p<0.05. (F) Quantification of the half-time (t1/2) of VDAC1 modification in cells from (C) over 6 hr. Half-times were obtained from decay curves generated with the time-points in (C). Bars represent mean ± SEM, n = 5 experiments. (G) Immunoblot analysis of U2OS:GFP-parkin cells, transfected with the indicated siRNA, treated with 20 μM CCCP in the presence or absence of 25 μM NMS-873 for six hours. (H) Immunoblot quantification of VDAC1 levels (relative to actin) in cells from (G), represented as a fold change in VDAC1 remaining when NMS-873 is added. Data points are represented on the graph, n = 3 experiments. Factors promoting ER-OMM contact are contained within the blue box.