Extended Data Figure 10. Model explaining cNCC-type selective effects of nucleolar dysfunction and rDNA damage in TCS.
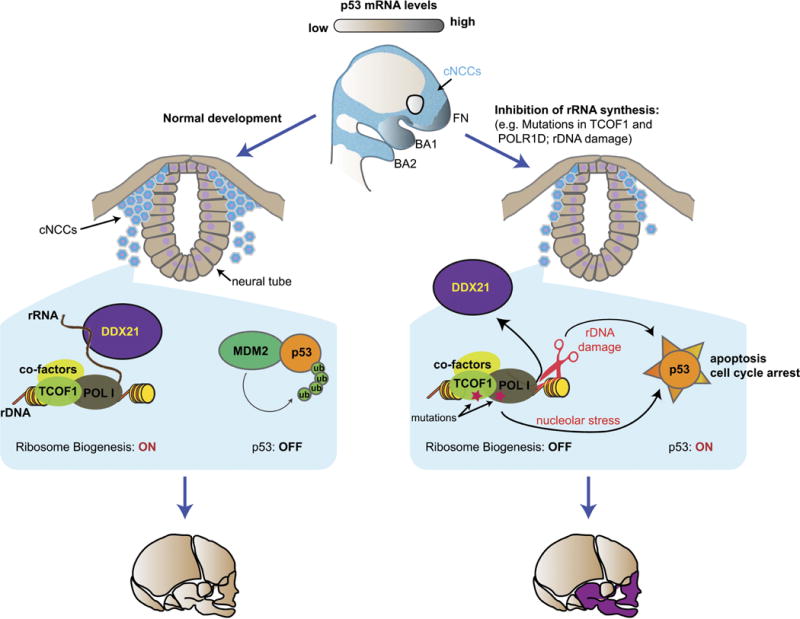
cNCCs express high levels of p53 mRNA, but during normal development p53 is under posttranscriptional control of its E3 ligase, Mdm2. Upon nucleolar stress and/or rDNA damage, activation of p53 and loss of DDX21 from chromatin result in apoptosis of a subset of cNCCs. This diminishes the population of cNCCs that can be allocated to the lower face, leading to malformations of the developing craniofacial structures. Thus, factors whose perturbations may ultimately induce defects in rRNA synthesis and rDNA damage are likely to be associated with craniofacial malformations.
