Extended Data Figure 3. Generation of an in vitro model of TCS.
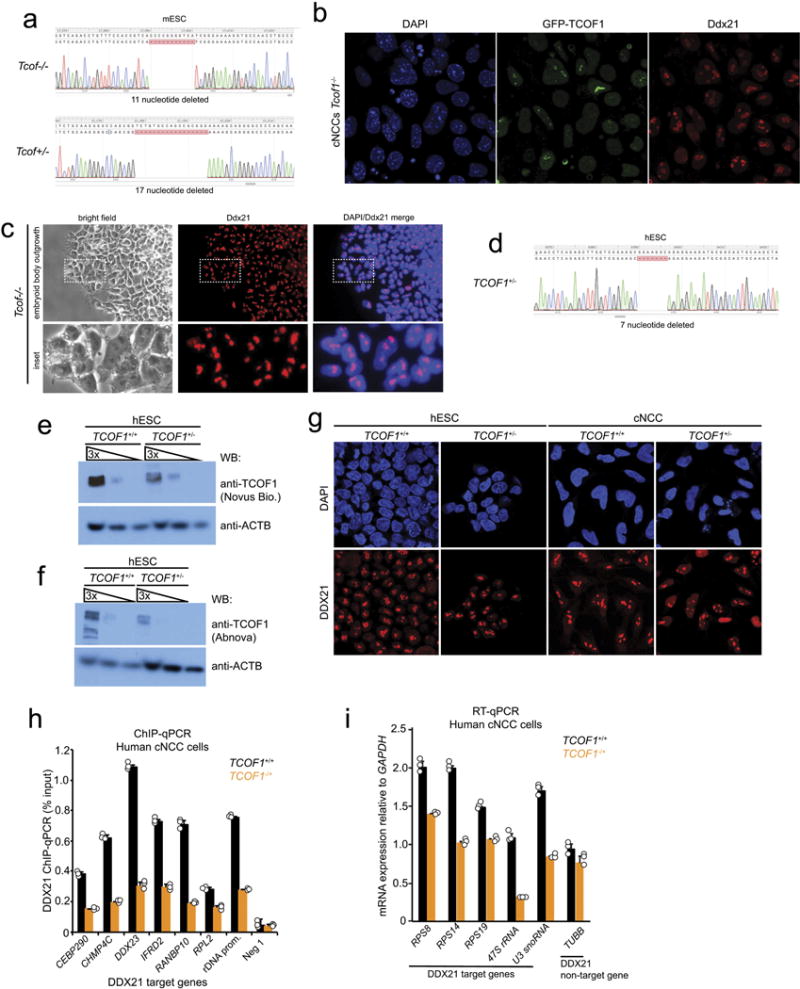
a, Mouse ES cells were co-transfected with CRISPR-Cas9 and sgRNAs targeting the Tcof1 locus. Targeted mouse ES cells were single-cell cloned and screened for loss-of-function mutations in Tcof1. Clones of the indicated genotypes were selected for this study. b, Overexpression of an exogenous, but stably integrated human GFP–tagged TCOF1 construct in mouse cNCCs rescued DDX21 localization defects as determined by immunofluorescence (quantifications are shown in Fig. 2c). Shown are representative images from n = 3 biologically independent experiments. c, Mouse ES cells were differentiated into embryoid bodies. Embryoid body outgrowth explants were further grown in culture and stained with antibodies for Ddx21. Shown are representative images from n = 4 biologically independent experiments. d, Human H9 ES cells were co-transfected with CRISPR-Cas9 and sgRNAs targeting the TCOF1 locus. Targeted ES cells were cloned and screened for loss-of-function mutations in TCOF1; unlike mouse ES cells, we did not recover homozygous mutant alleles for TCOF1 in human cells. The indicated genotype was selected for this study. e, f, Two different commercially available antibodies raised against TCOF1 were used to confirm the heterozygosity of TCOF1+/− ES cells. Shown are representative western blots from n = 2 biologically independent experiments. g, Representative immunofluorescence images showing DDX21 localization in both wild-type and TCOF1+/− human ES cells and cNCCs from n = 3 biologically independent experiments. h, ChIP–qPCR analysis in human cNCCs sampling DDX21 genomic occupancy at a representative panel of DDX21 target promoters and at the rDNA promoter. i, qPCR analyses of DDX21-regulated Pol I and Pol II transcribed ribosomal genes. For h, i, bars represent the average of n = 3 biologically independent experiments; error bars, s.e.m.
