Extended Data Figure 7. DDX21 overexpression rescues TCS and its function is deregulated by knockdown of other ribosomopathy-associated genes.
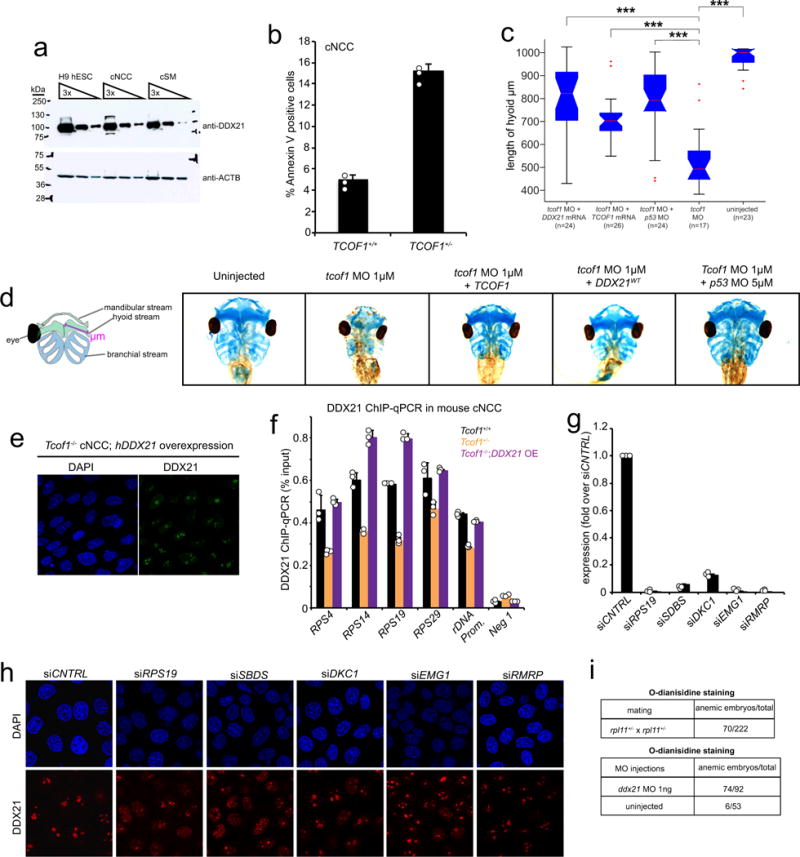
a, Representative western blot for DDX21 in different cell types from n = 3 biologically independent experiments. b, FACS analyses to determine the sensitivity of TCOF1+/− cNCC to p53-mediated apoptosis. cNCCs were treated with NSC146109 for 4–6 h (note that this time point is significantly shorter than the one used in Fig. 3f and g). Apoptosis was quantified by FACS of annexin V staining. Bars represent the average of n = 3 independent experiments; error bars, s.e.m. c, Quantification of Xenopus cranofacial development rescue experiments by measuring the length of the hyoid stream upon overexpression of TCOF1, DDX21, or p53 knockdown. Embryos were collected from n = 3 biologically independent experiments. Boxes represent median value and 25th and 75th percentiles, whiskers are minimum to maximum, crosses are outliers. ***P < 0.001, two-sided Wilcoxon–Mann–Whitney test. d, Rescue of TCS-associated craniofacial malformations in Xenopus by injection of the embryos with the indicated in vitro transcribed mRNAs and/or morpholinos (quantification is shown in c). e, Representative immunofluorescence images of mouse Tcof1−/− cNCCs upon overexpression of human GFP–tagged DDX21. n = 3 biologically independent experiments. f, ChIP–qPCR analysis, in mouse cNCCs sampling Ddx21 genomic occupancy, at a representative panel of Ddx21 target promoters and at the rDNA locus. n = 3 biologically independent experiments. Bars represent the average of n = 3 independent experiments; error bars, s.e.m. g, siRNA pools against a subset of ribosomopathy-associated genes were transected into HeLa cells. qPCR was used to determine the efficiency of the knockdowns. Bars represent the average of n = 3 independent experiments; error bars, s.e.m. h, Representative immunofluorescence images showing DDX21 localization changes in HeLa cells transfected with the indicated pools of siRNAs (quantification is on Fig. 3i). n = 3 biologically independent experiments. i, Tables quantifying the number of embryos stained for haemoglobin with o-dianisidine for the indicated genotypes. In the case of rpl1 zebrafish, embryos were collected from three independent matings. For DDX21, three independent batches of embryos were injected and stained.
