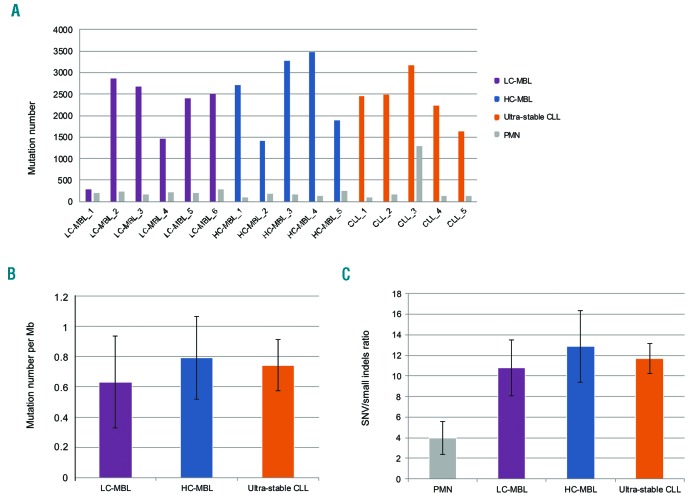
Figure 1.
Somatic mutational analysis of ultra-stable chronic lymphocytic leukemia (CLL), high-count monoclonal B-cell lymphocytosis (HC-MBL), low-count monoclonal B-cell lymphocytosis (LC-MBL) and control polymorphonuclear (PMN) cell samples. (A) Total number of somatic mutations identified by whole-genome sequencing (WGS) in CLL cell samples from MBL, CLL and the respective PMN samples. All samples carried similar mutational loads with the exception of a single LC-MBL sample (LC-MBL_1) that displayed a very low number of mutation events; as can be seen, the corresponding PMN sample had a mutation load similar to the other PMN samples, where comparison of mutation profiles between the MBL and PMN sample showed few common hits, thus excluding the likelihood of contamination. Concerning PMN control samples, they were also characterized by high homogeneity regarding the mutational load. There was a single sample with a very high mutational load; detailed comparison against its respective CLL sample showed a high overlap of mutations indicating potential tumor cell contamination, hence this sample was removed from downstream analysis. (B) Average mutation rates ± Standard Deviation (SD) for LC-MBL, HC-MBL and CLL. Highly analogous mutation rates were observed in the HC-MBL (0.79 mutations per Mb) and CLL (0.74 mutations per Mb) samples, while LC-MBL samples had a slightly lower ratio (0.63 mutations per Mb). (C) Average SNV to small indels ratio ± SD for all sample groups. All 3 entities displayed similar ratios in clear contrast to the PMN samples where the ratio was much lower.