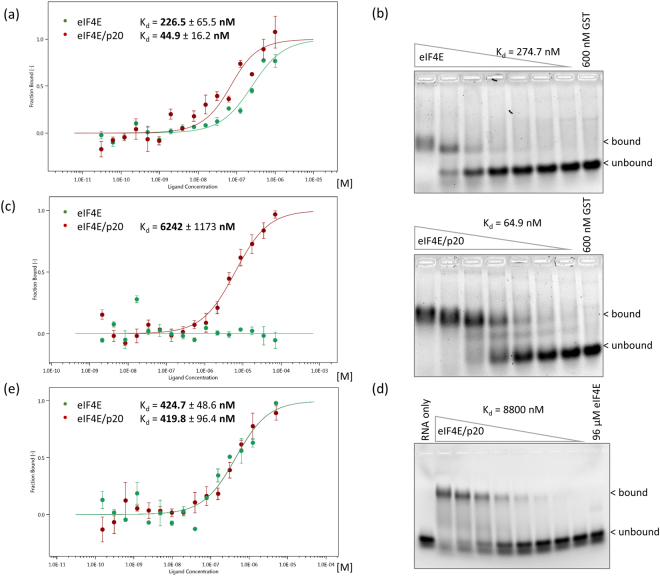
Figure 1.
Determination of dissociation constants (Kd) for eIF4E/p20 complex or eIF4E bound to capped or uncapped RNA by MicroScale Thermophoresis (MST) and Electrophoretic Mobility Shift Assay (EMSA). (a) MST: 50 nM RED fluorescence dye-labeled His6x-eIF4E (green) or eIF4E/His6x-p20 complex (red) with serially diluted m7GpppG-capped 64 nt long RNA (40 nt of SSA1 5′UTR), starting at 1 µM. Error bars indicate s.d. (n = 3). (b) EMSA: 600–0 nM His6x-eIF4E or 600–0 nM eIF4E/His6x-p20 complex with 0.25 µM m7GpppG-capped 64 nt long RNA (40 nt of SSA1 5′UTR) probe. SYBR Gold Nucleic Acid Gel Stain. GST (Glutathione-S Transferase) serves as a negative control. (c) MST: Serially diluted His6x-eIF4E (green) or eIF4E/His6x-p20 complex (red), starting at 96 µM, with 50 nM 3′FAM-labeled uncapped 40 nt long 5′UTR of SSA1 RNA. Error bars indicate s.d. (n = 3). (d) EMSA: 96–0 µM eIF4E/His6x-p20 complex with 1 µM 3′FAM-labeled uncapped 40 nt long RNA probe (5′UTR of SSA1). (e) MST: 50 nM RED fluorescence dye-labeled His6x-eIF4E (green) or eIF4E/His6x-p20 complex (red) with serial dilutions of m7GpppG starting at 5 µM. Error bars indicate s.d. (n = 3).