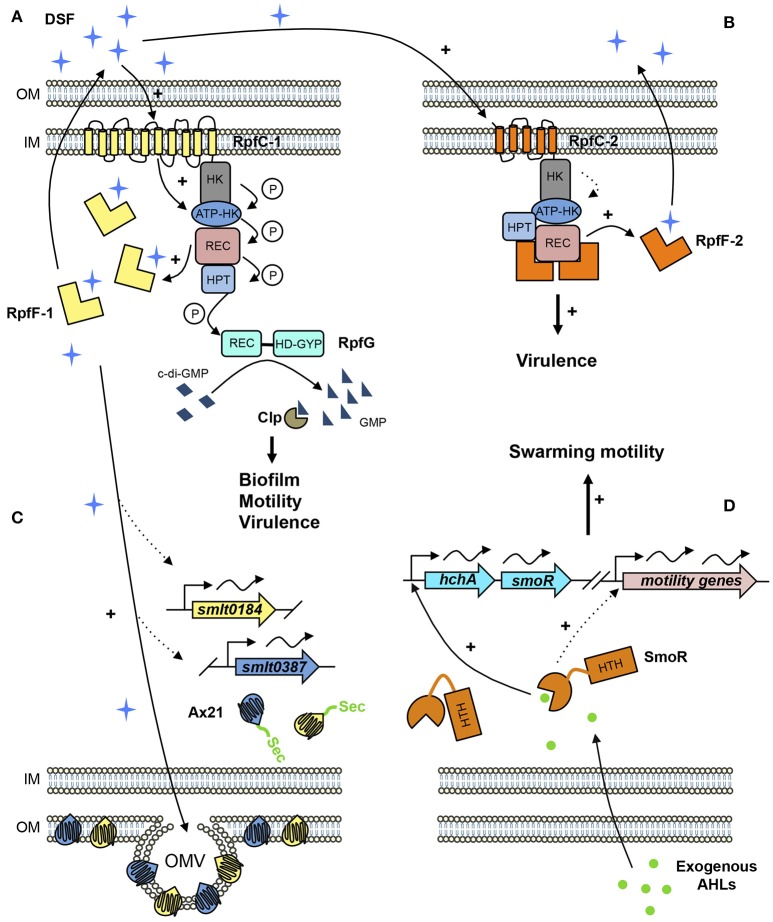
Figure 1.
Proposed QS signaling network in S. maltophilia. (A) In rpf-1 strains, RpfC-1 (including 10 TMR) stimulates RpfF-1 basal activity—that increases with cell density—and synthesizes DSF (cis-11-Methyl-2-dodecenoic acid) that accumulates in the extracellular environment. Once DSF concentration reaches a critical threshold, RpfC-1 senses DSF, and induces a phosphorylation cascade throughout its cytoplasmic domains ending in the response regulator RpfG, which degrades cyclic diguanylate monophosphate (c-di-GMP) to GMP activating the transcriptional regulator Clp that stimulates expression of genes involved in biofilm formation, motility, and virulence. (B) In rpf-2 strains, RpfC-2 (5 TMR) permanently represses RpfF-2, resulting in no DSF detection in axenic conditions. DSF produced by neighboring bacteria (e.g., rpf-1 strain) is sensed by RpfC-2 allowing free-active RpfF-2 and subsequent DSF synthesis. (C) DSF also stimulates the production of outer membrane vesicles (OMV) containing high amounts of the two Ax21 proteins (Smlt0184 and Smlt0387). Both Ax21 proteins present a signal peptide that is processed by the general secretory (Sec) system. (D) Exogenous AHL signals, specifically C8-HSL and oxo-C8-HSL, are sensed by the LuxR solo SmoR (Smlt1839), annotated as “LuxR chaperone HchA-associated,” activating the transcription of its own operon and promoting swarming motility. Dotted lines represent predicted or supposed interactions on the basis of reported experimental evidences. Protein domains are abbreviated as follows. HK, Histidine kinase domain; REC, Receiver domain; HPT, Histidine phosphotransferase domain; HD-GYP, Phosphodiesterase domain containing an additional GYP motif; HTH, Helix-Turn-Helix domain.