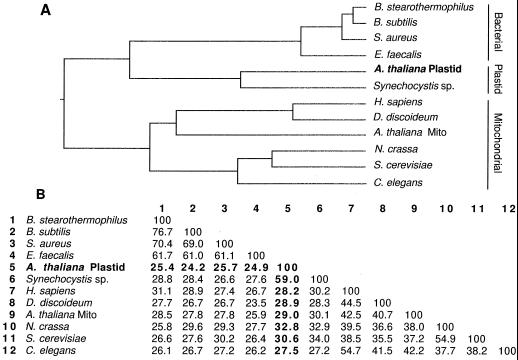
Figure 7.
Comparison of amino acid sequences of 12 E2s. A, The length of each branch of the dendrogram is proportional to sequence identity. The cDNA clone described here is in boldface (A. thaliana Plastid). The sequences segregate into three groups denoted on the right as Bacterial, Plastid, and Mitochondrial. Mito, Mitochondria. B, Amino acid identity matrix for the sequences in A. The identities of the plastid sequence with the other E2 sequences are in boldface. GenBank accession numbers for the sequences not included in Figure 6 are as follows: Bacillus stearothermophilus, X53560; Staphylococcus aureus, X58434; Dictyostelium discoideum, U06634; Neurospora crassa, J04432; and Caenorhabditus elegans, Z77659. The Enterococcus faecalis sequence can be accessed in the PIR database, accession no. S16989.