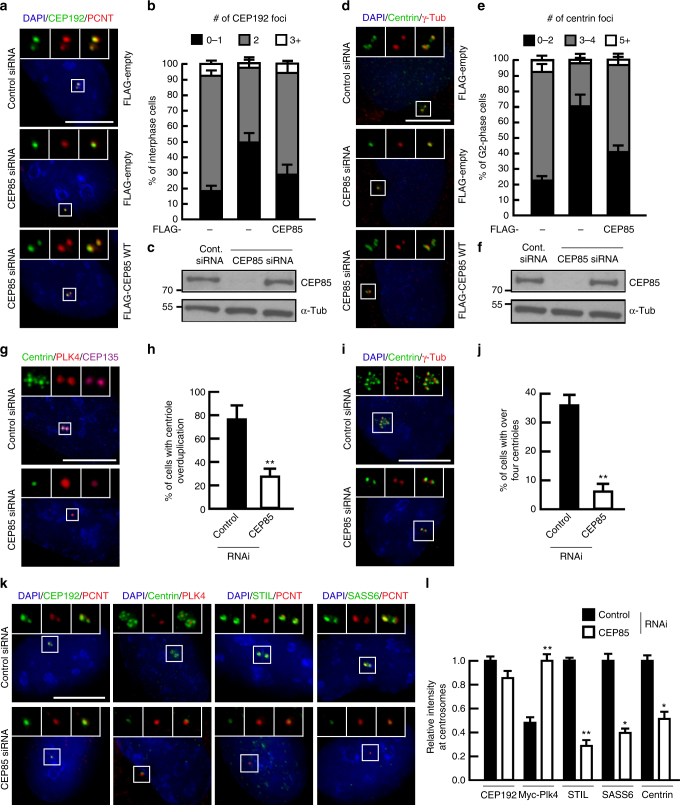
Fig. 1.
CEP85 is a regulator of centriole duplication. a–c Effect of CEP85 depletion on centrosome number. U-2 OS cells expressing Tet-inducible FLAG or the siRNA-resistant FLAG-CEP85 transgene were transfected with control or CEP85 siRNA and induced with tetracycline for 72 h. Scale bar 10 μm, white boxes indicate the magnified region. b The graph shows the percentage of cells with the indicated centrosome numbers (n = 200/experiment, three independent experiments). c Western blot analysis of FLAG-CEP85 protein levels. α-tubulin served as a loading control. d–f Impact of CEP85 depletion on centriole number. The G2-phase arrest assays (see Methods) were performed in U-2 OS cells conditionally expressing FLAG or siRNA-resistant FLAG-CEP85, treated with control or CEP85 siRNA and induced with tetracycline for 72 h. Scale bar 10 μm, white boxes indicate the magnified region. e Bar graph, the number of centrioles per cell were counted. (n = 200/experiment, three independent experiments). f Western blot showing the levels of FLAG-CEP85 in control or CEP85 siRNA transfected cells. α-tubulin served as a loading control. g, h The role of CEP85 in PLK4-induced centriole overduplication. The PLK4 assays were performed as described in Methods. Scale bar 10 μm, white boxes indicate the magnified region. h The graph showing the percentage of cells with over four centrioles (n = 100/experiment, three independent experiments). i, j The role of CEP85 in S-phase arrest induced centriole overduplication. The assays were performed as described in Methods. Scale bar 10 μm, white boxes indicate the magnified region. j The graph indicates the percentage of cells with over four centrioles (n = 100/experiment, three independent experiments). k, l Determining at which stage CEP85 acts in the centriole duplication pathway using the PLK4-induced centriole overduplication assays (see the Methods). Scale bar 10 μm, white boxes indicate the magnified region. l The graph indicates the relative levels of CEP192, Myc-Plk4, STIL, SASS6 and Centrin at centrosomes (n = 100/experiment, three independent experiments). Two-tailed t-test was performed for all p-values, all error bars represent SD, and asterisks for p-values are **p < 0.01 and *p < 0.05