Figure 5.
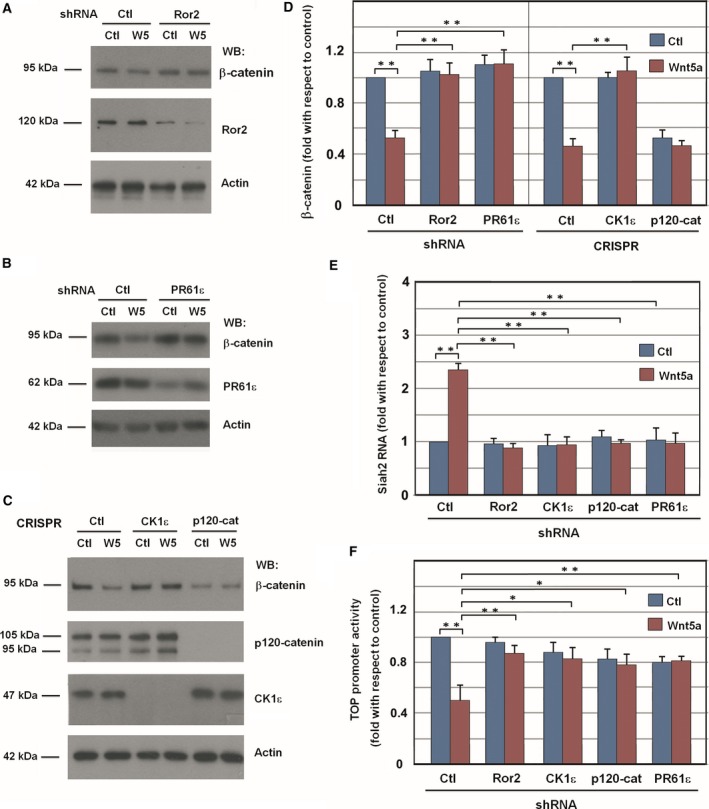
Downregulation of Ror2, p120‐catenin, CK1ε, or PR61ε prevents the β‐catenin down‐modulation induced by Wnt5a. HEK293T cells were depleted of Ror2 (A) or PR61ε (B) using specific shRNA; a scrambled shRNA was used as a control. After 48 h, cells were stimulated with control or Wnt5a‐conditioned medium overnight, and β‐catenin levels were analyzed by WB from total cell extracts. (C) Control, p120‐catenin, and CK1ε CRISPR HEK293T cells were treated with control or Wnt5a‐conditioned medium overnight, and β‐catenin was analyzed by WB. (D) β‐catenin levels were quantified by analyzing three independent experiments performed in (A–C) (mean ± SD). **P < 0.01. (E) RNA was isolated from control, Ror2, CK1ε, p120‐catenin, and PR61ε‐depleted HEK293T cells stimulated overnight with control or Wnt5a‐conditioned medium. Expression of SIAH2 was assessed by semi‐quantitative RT‐PCR. Results are presented as mean ± SD from three independent experiments. *P < 0.05; **P < 0.01. (F) β‐catenin transcriptional activity was determined using the TOP‐Flash reporter plasmid in SW‐480 cells transfected with the indicated shRNA. pTK‐Renilla plasmid was transfected to normalize the efficiency of transfection. Relative luciferase activity was determined 48 h after transfection. Cells were treated with Wnt5a for 16 h before cell lysis. FOP‐Flash plasmid was also transfected as a control; the activity of this promoter was always lower than 1% of the value of obtained with TOP‐Flash. The mean ± SD of four experiments is shown. **P < 0.01.
