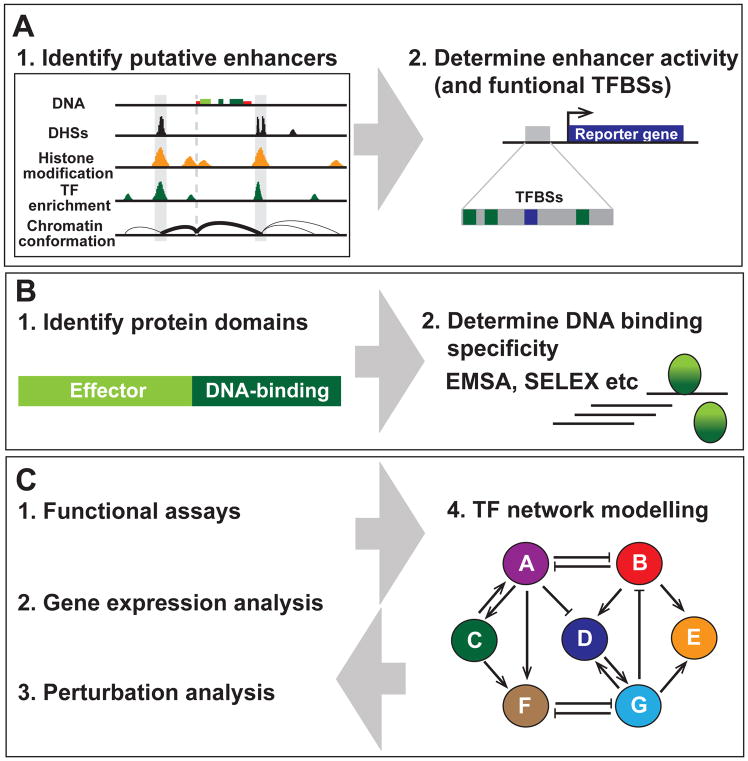
Figure 2. Approaches to build TF regulatory network models.
(A) Enhancers. Putative enhancers can be identified by a number features including DNase I hypersensitivity sites (DHSs), histone modifications (such as H3K4me), TF enrichment and DNA looping (measured by chromatin conformation capture methods such as Hi-C). Enhancer activity can be assessed using in vivo or in vitro enhancer assays, and the function of the TFBSs (DNA motifs) identified within such enhancers can assessed by mutational analysis.
(B) Transcription factors. TFs can be identified by their DNA binding domains. TFs also contain effector domains, which are responsible for protein-protein interactions. A range of methods including electrophoretic mobility shift assays (EMSAs) and systematic evolution of ligands by exponential enrichment (SELEX) have been used to determine individual and cooperative TF DNA binding specificities.
(C) Building TF network models. Methods in (A) and (B) can be combined with functional assays (such as enhancer mutagenesis), gene expression analysis and/or TF pertubation analysis to build and train TF regulatory network models that can be “executed” in silico. These models provide important insights into the biological logic underpinning mammalian cell fate decisions, which feedback into experimental research.