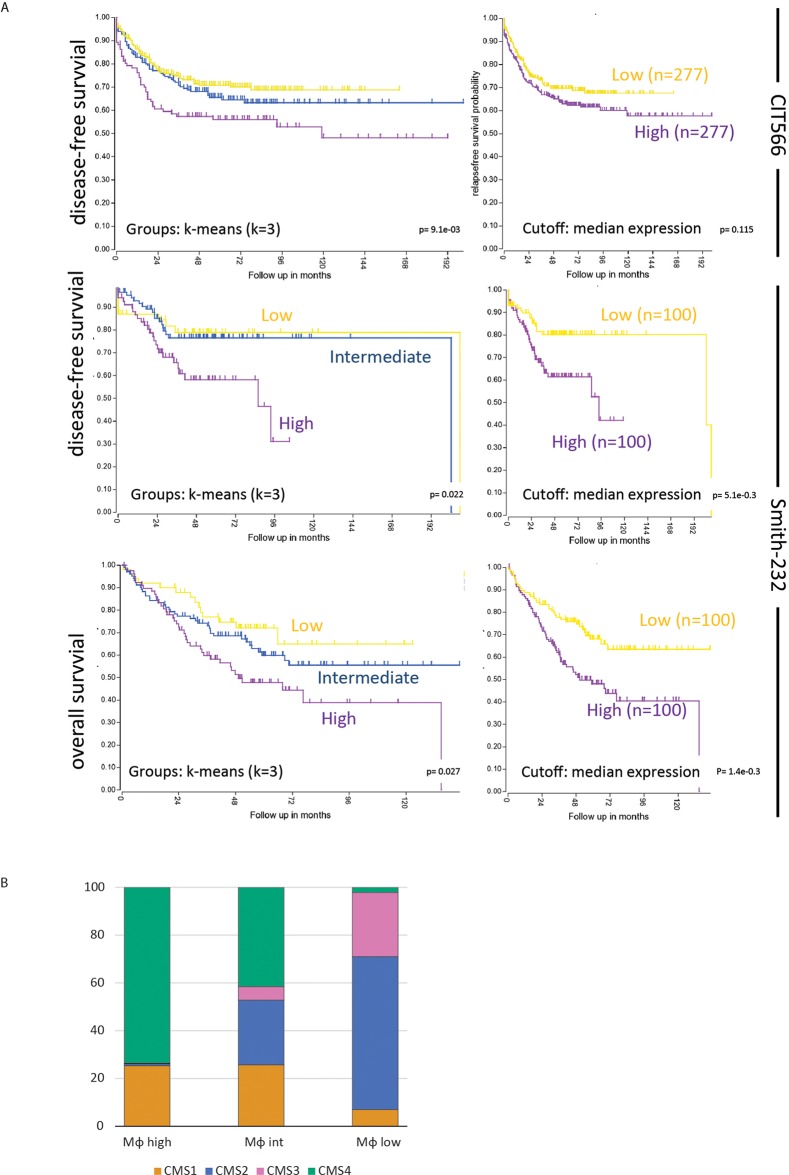
Figure 1. A macrophage signature identifies a poor prognosis subgroup of primary human colorectal tumors.
(A) The macrophage signature of the immune compendium was used to cluster the tumors of the CIT566 cohort into three groups by k-means clustering (http://r2.amc.nl): Macrophage high (n=142), intermediate (n=264) and low (n=396). The Kaplan Meier curve shows the differences in disease-free survival of the three subgroups. The inset shows the signature expression levels in the three subgroups. Moreover, by generating single meta-gene values of the multi-gene macrophage signature and taking the median expression of those meta-gene values as a cutoff, two equally sized groups of tumors were created in both the CIT566 and Smith cohorts. These tumors showed significant differences in disease-free and overall survival, similar to the k-means clustering. Characteristics of these cohorts shown in Supplementary Figure 1C. (B) Bar graphs showing the contribution of the four CMS subtypes to macrophage-high, -intermediate and -low subgroups in the CMS-3232 cohort. The CMS classification was derived from Guinney et al. [9].