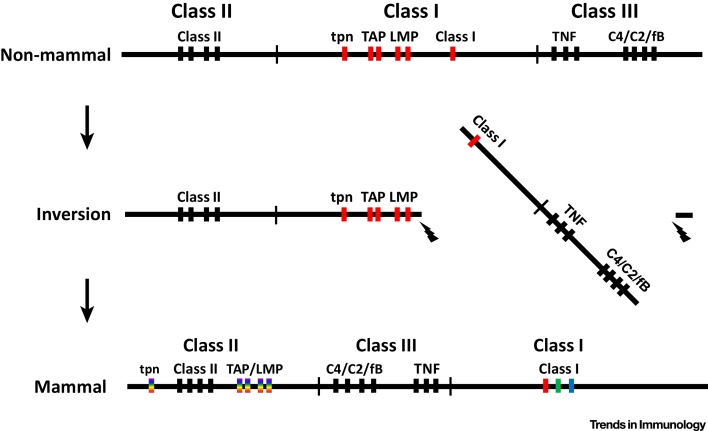
Figure 3.
The Presence of a Multigene Family of Well-Expressed Classical Class I Molecules in Typical Placental Mammals Can Be Explained by a Genomic Inversion that Disrupted the Coevolutionary Relationships between the Closely Linked Genes of the Class I System Found in Many Other Vertebrates. Top panel. The genomic organization of an ancestral major histocompatibility complex (MHC) haplotype, based on data from the chicken and throughout the non-mammalian vertebrates, has class II genes in a class II region, class I genes and the genes encoding antigen-processing and peptide-loading components in a class I region, and the class III region genes on the outside. The close linkage within the class I region leads to a single dominantly expressed class I gene (red) whose peptide motif reflects the specificities of the polymorphic antigen-processing and peptide-loading genes (all red) with which it coevolves. Middle panel. A genomic inversion can lead to the class III region moving between the single dominantly expressed class I gene and the rest of the MHC, marooning the particular alleles of the antigen-processing and peptide-loading genes near the class II genes and far from the class I allele that they serve. Bottom panel. The antigen-processing and peptide-loading genes are selected to support any class I allele that might appear due to recombination (rainbow color), which would then allow duplication within an MHC haplotype to give a multigene family encoding class I molecules with different peptide motifs (red, green, blue), as is found in typical mammals. Regions separated by thin vertical lines; genes indicated by thicker vertical lines; tpn, tapasin; TAP, transporter associated with antigen presentation; LMP, inducible proteasome component, originally known as low molecular weight protein; C2, complement component 2; C4, complement component 4; fB, factor B. Modified from [9].