ABSTRACT
Bifidobacterial carbohydrate metabolism has been studied in considerable detail for a variety of both plant- and human-derived glycans, particularly involving the bifidobacterial prototype strain Bifidobacterium breve UCC2003. We recently elucidated the metabolic pathways by which the human milk oligosaccharide (HMO) constituents lacto-N-tetraose (LNT), lacto-N-neotetraose (LNnT) and lacto-N-biose (LNB) are utilized by B. breve UCC2003. However, to date, no work has been carried out on the regulatory mechanisms that control the expression of the genetic loci involved in these HMO metabolic pathways. In this study, we describe the characterization of three transcriptional regulators and the corresponding operator and associated (inducible) promoter sequences, with the latter governing the transcription of the genetic elements involved in LN(n)T/LNB metabolism. The activity of these regulators is dependent on the release of specific monosaccharides, which are believed to act as allosteric effectors and which are derived from the corresponding HMOs targeted by the particular locus.
IMPORTANCE Human milk oligosaccharides (HMOs) are a key factor in the development of the breastfed-infant microbiota. They function as prebiotics, selecting for a specific range of microbes, including a number of infant-associated species of bifidobacteria, which are thought to provide a range of health benefits to the infant host. While much research has been carried out on elucidating the mechanisms of HMO metabolism in infant-associated bifidobacteria, to date there is very little understanding of the transcriptional regulation of these pathways. This study reveals a multicomponent transcriptional regulation system that controls the recently identified pathways of HMO metabolism in the infant-associated Bifidobacterium breve prototype strain UCC2003. This not only provides insight into the regulatory mechanisms present in other infant-associated bifidobacteria but also provides an example of a network of sequential steps regulating microbial carbohydrate metabolism.
KEYWORDS: bifidobacteria, probiotic, prebiotic, transcriptional regulation, HMO, carbohydrate metabolism
INTRODUCTION
Bifidobacteria represent high-G+C, Gram-positive, anaerobic members of the phylum Actinobacteria and are common commensals of the mammalian, avian, and insect guts. In humans, they are particularly abundant and prevalent among the gut microbiota of healthy, vaginally delivered, breastfed infants (1) and are thought to confer a multitude of benefits to the neonatal host (2–4). For this reason, as well as because of their purported health-promoting activities in adults, bifidobacteria are used as functional ingredients in a variety of foods and therapeutic products. The use of prebiotics is also becoming commonplace for the improvement of both adult and infant (gut) health. A prebiotic has been defined as “a nondigestible food ingredient that beneficially affects the host by selectively stimulating the growth and/or activity of one or a limited number of bacteria in the colon, and thus improves host health” (5).
Fascinatingly, the archetypal prebiotic would appear to be human breast milk, in particular its bifidogenic constituents known as human milk oligosaccharides (HMOs). HMOs represent specific glycans present in human breast milk that are thought to shape, at least partly, the compositional structure of the neonatal gut microbiota (6, 7). HMOs represent, after lactose, the second-largest carbohydrate component of breast milk (6, 8) and constitute a heterogeneous mix of at least 200 distinct glycan structures (9). The majority of complex HMO structures can be classified into one of two types, depending on their backbone composition. The more abundant type I HMOs contain the core tetrasaccharide lacto-N-tetraose (LNT) within their structure (Galβ1-3GlcNAcβ1-3Galβ1-4Glc). Type II HMOs contain lacto-N-neotetraose (LNnT), a stereoisomer of LNT, within their backbone (Galβ1-4GlcNAcβ1-3Galβ1-4Glc). Lacto-N-biose (LNB) (Galβ1-3GlcNAc) is a subunit of LNT and other type I HMO structures and can be released by the degradation of these sugars (10).
The effects of specific human breast milk components on the prevalence, abundance, and activity of members of the infant gut microbiota are currently enjoying a great deal of scientific and commercial attention due to the beneficial roles that they are believed to play in infant health and development (11, 12). Understanding the pathways by which specific HMOs are metabolized by particular microbial species that inhabit the infant gut is important, although our knowledge regarding these processes is still in its infancy, particularly with regard to the manner in which they affect microbiota development.
It is hardly surprising that the dominant Bifidobacterium species found among the neonatal gut microbiota can utilize various HMO components as their sole carbohydrate source (9). These species chiefly include strains of Bifidobacterium bifidum, Bifidobacterium longum subsp. infantis, and Bifidobacterium breve. HMO utilization by B. bifidum and B. longum subsp. infantis is relatively well characterized. B. bifidum extracellularly hydrolyzes complex HMO structures, including LNT and LNnT, by employing secreted glycosyl hydrolases, followed by the internalization and intracellular degradation/metabolism of (most of) the resulting mono- and disaccharides, such as LNB (10, 13–19). B. longum subsp. infantis internalizes intact LNT, LNnT, and LNB and uses a series of sequential hydrolytic/phosphorolytic reactions acting from the nonreducing end of the carbohydrate structures to degrade them into their monosaccharide components for further metabolic processing (9, 10, 20–23). However, B. infantis has also been demonstrated to take up and utilize fucosyl- and sialyl-lactose (24–26).
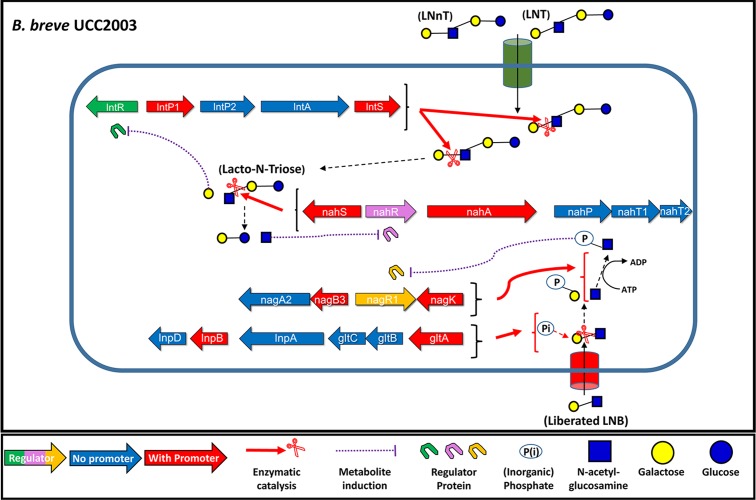
The metabolic pathways of LNT, LNnT, and LNB have recently been elucidated for the prototype strain B. breve UCC2003 (27). In that study, converging pathways of LNT and LNnT catabolism were identified, where monosaccharide moieties are sequentially released from the nonreducing end of either sugar by hydrolytic reactions. The genetic units responsible for the uptake and breakdown of these structures are the lnt locus (corresponding to locus tags Bbr_0526 to Bbr_0530) and the nah locus (locus tags Bbr_1554 to Bbr_1560) (Table 1 and Fig. 1). The lnt locus encodes proteins that are responsible for the internalization of LNT and the intracellular hydrolysis of both LNT and LNnT, releasing a galactose (Gal) moiety from their nonreducing end and at the same time liberating the trisaccharide lacto-N-triose (GlcNAcβ1-3Galβ1-4Glc). The nah locus specifies an LNT/LNnT uptake system, while it furthermore encodes a glycosyl hydrolase that liberates N-acetylglucosamine (GlcNAc) from the nonreducing end of lacto-N-triose, leaving lactose, which itself is further degraded by lactose-specific glycosyl hydrolases. Additionally, the gene products of the lnp-glt locus (corresponding to locus tags Bbr_1585 to Bbr_1590) (Table 1 and Fig. 1) are responsible for the internalization and subsequent phosphorolysis of free LNB, releasing its constituent monosaccharides Gal-1-phosphate and GlcNAc (27, 28). We also identified the transcriptional upregulation of genes in the nag locus (locus tags Bbr_1247 to Bbr_1252) (Table 1 and Fig. 1) during growth on LNT, LNnT, and LNB, indicating their role in the utilization of these sugars, specifically in the multistep metabolism of GlcNAc. The nag locus was previously implicated in the metabolism of sialic acid and mucin-derived N-glycans, both of which contain GlcNAc as well (29, 30). While the degradation routes of these key HMO structures have thus been identified, the regulatory mechanisms that control the expression of these pathways have remained unexplored, for both B. breve and HMO-utilizing Bifidobacterium species as a whole.
TABLE 1.
B. breve UCC2003 regulator mutant genes with upregulated transcription during growth in mMRS medium supplemented with 1% ribose as the sole carbohydrate, compared to the wild type (control)
| Locus tag | Gene | Function | Fold upregulationa during growth of: |
||
|---|---|---|---|---|---|
| UCC2003-lntR | UCC2003-nahR | UCC2003-nagR1 | |||
| Bbr_0526 | lntR | Transcriptional regulator, LacI family | NA | — | — |
| Bbr_0527 | lntP1 | Permease protein of the ABC transporter system for sugars | 3.84 | — | — |
| Bbr_0528 | lntP2 | Permease protein of the ABC transporter system for sugars | 3.77 | — | — |
| Bbr_0529 | lntA | GH42 beta-galactosidase | 2.78 | — | — |
| Bbr_0530 | lntS | Solute-binding protein of the ABC transporter system for sugars | 5.66 | — | — |
| Bbr_1247 | nagA2 | CE9 nagA2 N-acetylglucosamine-6-phosphate deacetylase | — | — | 6.70 |
| Bbr_1248 | nagB3 | nagB3 glucosamine-6-phosphate isomerase | — | — | 9.11 |
| Bbr_1249 | nagR1 | Transcriptional regulator, ROK family | — | — | NA |
| Bbr_1250 | nagK | Sugar kinase, ROK family | — | — | 2.29 |
| Bbr_1251 | nagR2 | Transcriptional regulator, ROK family | — | — | — |
| Bbr_1252 | nagK2 | Sugar kinase, pfkB family | — | — | — |
| Bbr_1554 | nahS | Solute-binding protein of the ABC transporter system (lactose) | — | 17.44 | — |
| Bbr_1555 | nahR | NagC/XylR-type transcriptional regulator | — | NA | — |
| Bbr_1556 | nahA | GH20 nagZ beta-N-acetylhexosaminidase | — | — | — |
| Bbr_1558 | nahP | Permease protein of the ABC transporter system | — | — | — |
| Bbr_1559 | nahT1 | ATP-binding protein of the ABC transporter system | — | — | — |
| Bbr_1560 | nahT2 | ATP-binding protein of the ABC transporter system | — | — | — |
| Bbr_1585 | lnpD | UDP-glucose 4-epimerase | — | — | 3.06 |
| Bbr_1586 | lnpB | Phosphotransferase family protein | — | — | 3.36 |
| Bbr_1587 | lnpA | GH112 lacto-N-biose phorylase | — | — | 2.90 |
| Bbr_1588 | gltC | Permease protein of the ABC transporter system for sugars | — | — | 2.91 |
| Bbr_1589 | gltB | Permease protein of the ABC transporter system for sugars | — | — | 3.02 |
| Bbr_1590 | gltA | Solute-binding protein of the ABC transporter system for sugars | — | — | 5.07 |
The levels of transcription are shown as fold increases in transcription levels on each carbohydrate, compared to a ribose control. Data are based on comparative transcriptome analysis using B. breve UCC2003-lntR, B. breve UCC2003-nahR, and B. breve UCC2003-nagR1 grown on 1% ribose, compared to wild-type B. breve UCC2003 grown under the same conditions as a control. Two independent biological replicates were used for each array, using a Cy3/Cy5 dye swap. The cutoff point is 2.0-fold, with a P value of 0.001. —, value below the cutoff. NA indicates that the fold increase in the transcription level of this gene is not included, as this is the gene in which the mutation was made, and thus, it does not accurately represent its natural transcription under these conditions. The level of transcription is not given for the regulator-encoding genes containing the mutations in their respective arrays, as their transcription has been interrupted and thus cannot be considered reliable.
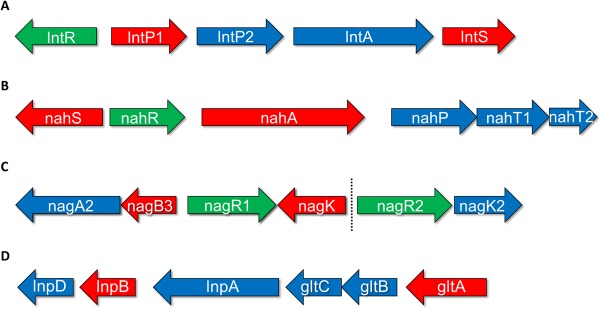
FIG 1.
Schematic representation of HMO metabolism-associated loci in B. breve UCC2003, as identified previously (21). (A) The genes of the lnt locus; (B) the genes of the nah locus; (C) the genes of the nag locus and the adjacent genes nagR2 and nagK2; (D) the genes of the lnp-glt locus. The length of the arrows is proportional to the size of the open reading frame. Genes shown in red possess a predicted promoter in their upstream intergenic region. Genes shown in green are predicted to encode a regulator protein. Genes shown in blue were identified as not possessing a predicted promoter in their upstream intergenic region.
In this study, we identified and characterized the genes encoding transcriptional regulators responsible for the control of gene expression in four key HMO-associated loci in B. breve UCC2003 during growth on LNT, LNnT, or LNB.
RESULTS
Identification of putative transcriptional regulator-encoding genes in the vicinity of HMO utilization loci.
In a previous study, we observed that genes within four chromosomal loci exhibit transcriptional induction during the growth of B. breve UCC2003 on LNT, LNnT, or LNB as the sole carbohydrate source (27). This indicates that these genes are subject to transcriptional regulation, which was presumed to be either directly or indirectly controlled by the presence of these HMO substrates. The four loci concerned are the lnt locus (Bbr_0526 to Bbr_530), the nah locus (Bbr_1554 to Bbr_1560), the nag locus (Bbr_1247 to Bbr_1250), and the lnp-glt locus (Bbr_1585 to Bbr_1590) (Fig. 1; see Table 1 for a description of [predicted] functions). Detailed scrutiny of these four loci and neighboring regions showed that the lnt and nah loci are flanked by or contain a predicted regulator-encoding gene, respectively: lntR (Bbr_0526), encoding a LacI-type repressor, and nahR (Bbr_1555), encoding a NagC/XylR-type repressor (Fig. 1A and B). The nag locus is associated with two genes, nagR1 (Bbr_1249) and nagR2 (Bbr_1251), both of which are predicted to encode repressor open reading frame kinase (ROK)/NagC family-type repressors, while no regulator-encoding gene was observed in the close vicinity of the lnp-glt locus (Fig. 1C and D). NagC/XylR-type and ROK/NagC-type repressors are both members of the large family of ROK-type transcriptional regulators (31). The four identified putative regulator-encoding genes were thus selected as candidates for mutagenesis in order to ascertain their role, if any, in the transcriptional regulation of the lnt, nah, nag, and lnp-glt loci.
Generation and transcriptomic analysis of insertional mutants in putative HMO-associated regulator-encoding genes.
Individual insertional mutants were constructed in lntR, nahR, nagR1, and nagR2, resulting in B. breve strains UCC2003-lntR, UCC2003-nahR, UCC2003-nagR1, and UCC2003-nagR2, respectively (see Materials and Methods). In order to identify promoters/genes that are subject to the transcriptional control of these predicted regulators, global gene transcription data were obtained from microarray-based analyses performed on the B. breve UCC2003-lntR, UCC2003-nahR, UCC2003-nagR1, and UCC2003-nagR2 insertion mutants grown in modified de Man-Rogosa-Sharpe (mMRS) medium supplemented with ribose and compared to the transcriptome of the UCC2003 wild-type strain grown under the same conditions.
Transcriptome analysis of the lntR mutant revealed the upregulation of the adjacent lntP1, lntP2, lntA, and lntS genes of the lnt locus (Table 1 and Fig. 1), when this mutant was grown on ribose (compared to wild-type UCC2003), all of which were also previously found to be upregulated during the growth of wild-type UCC2003 on LNT or LNnT (27). This corroborates the notion that LntR is a LacI-type repressor and that this protein negatively regulates the LNT/LNnT-dependent transcription of genes within the lnt cluster. Conversely, the array data obtained for the nahR mutant only revealed the transcriptional upregulation (compared to the UCC2003 control) of the nahS gene (Table 1), when grown on ribose. This is consistent with previously observed expression patterns for UCC2003, with the exception of nahA, which may have been expected to exhibit transcriptional upregulation in the nahR mutant, as its expression was increased during growth on LNT and LNnT in B. breve UCC2003 (27). These results suggest that NahR, a NagC/XylR-type repressor, is responsible for the transcriptional regulation of at least one gene of the nah cluster. For the nagR1 mutant, upregulation of nagA2, nagB3, and nagK (but not nagR2 or nagK2), as well as all of the genes of the lnp-glt locus (Table 1), was observed when cells were grown on ribose (compared to the UCC2003 control). These results suggest that NagR1, a ROK/NagC family-type repressor, is responsible for the transcriptional regulation of (part of) the nag and lnp-glt clusters. This is consistent with transcriptomic data previously obtained for wild-type UCC2003 during growth on LN(n)T and LNB, which demonstrated the transcriptional upregulation of genes in both of these loci (27). When the transcriptome of UCC2003-nagR2 was compared to that of UCC2003 grown on ribose, the nagR2 mutant exhibited increased transcription levels of genes in the mal locus (locus tags Bbr_0118 to Bbr_0123), which is known to be involved in maltooligosaccharide metabolism (32, 33), and Bbr_1719 to Bbr_1721 (predicted to function in fatty acid metabolism) (34) (data not shown), none of which are predicted to function in HMO metabolism, nor were they shown to be upregulated in our previous wild-type arrays for growth on LNT, LNnT, or LNB (27). These results thus show that NagR2 is not involved in the transcriptional control of the loci responsible for LNT, LNnT, or LNB metabolism, and no further investigation of this regulator was carried out. The lntR, nahR, and nagR1 genes, however, were selected for further study, as described below, in order to further elucidate their regulatory activities and specificities.
Promoter mapping through identification of transcription start sites.
Based on the transcriptome findings, we presumed that LntR, NahR, and NagR1 act as transcriptional regulators of (certain genes of) the lnt, nah, and nag-lnp-glt loci, respectively. Gene expression patterns observed for the regulator gene mutants and examination of the genetic layout and transcriptome profiles of these loci allowed us to assign putative promoter-containing regions within each locus. In order to verify these predicted promoter regions, the associated transcription start sites (TSSs) were experimentally determined by primer extension analyses.
The lnt locus was deduced to contain at least two promoters: one just upstream of lntP1 (Fig. 2a) and one in front of lntS (Fig. 2b). The lntP1 and lntS genes on the B. breve UCC2003 genome encode a permease and a solute-binding protein of an ABC transporter system, respectively, and exhibit increased transcription upon growth on LNT, LNnT, LNB, lactosamine, or lactose (27). The TSSs of the presumed lntP1 and lntS promoters were determined by primer extension analysis using RNA extracted from B. breve UCC2003 grown in mMRS medium supplemented with 1% LNnT. An extension product was identified 41 nucleotides 5′ of the predicted translational start site of the lntP1 gene (see Fig. S1A in the supplemental material), while the TSS for the lntS gene was identified 154 nucleotides 5′ of the predicted translational start site (Fig. S1B). In both cases, the TSS was preceded by −10 and −35 hexamers that resemble (bifidobacterial) consensus vegetative promoter recognition sequences (35, 36).
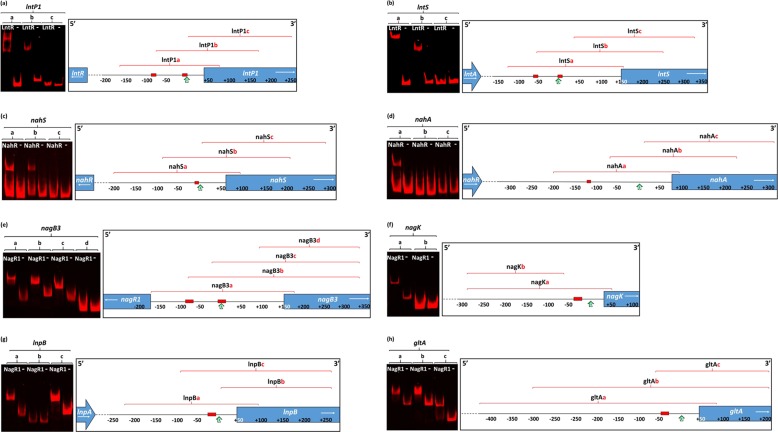
FIG 2.
EMSA images showing LntR (a and b), NahR (c and d), and NagR1 (e to h) interactions with a range of DNA fragments from the regions in the proximity of their predicted target promoters, in order to identify their approximate locations. The locations and sizes of fragments used, in relation to the promoter regions' respective transcription start sites, are given in Table S1 in the supplemental material. The panels at the right schematically represent the locations of the DNA fragments used in relation to the locations of the putative operator sequences (red boxes), transcription start sites (green arrows), and genes (arrows in blue boxes). In each panel, “−” indicates a negative control, where an equivalent amount of the crude cell extract from NZ9000 harboring empty plasmid pNZ8150 was added instead of the crude extract from the regulator-expressing NZ9000 strain.
The nah locus was deduced to contain at least two promoters: one just upstream of nahS (Fig. 2c) and one in front of nahA (Fig. 2d). The nahS and nahA genes on the B. breve UCC2003 genome encode a solute-binding protein of an ABC transporter system and a GH20 N-acetylhexosaminidase, respectively. While an increase in transcription was observed only for nahS in the nahR mutant-based array, both this gene and nahA were found to be subject to transcriptional induction when wild-type UCC2003 was grown on LNT, LNnT, or lactosamine (27). The TSSs of the presumed nahS and nahA promoters were determined by primer extension analysis using RNA extracted from B. breve UCC2003 grown in mMRS medium supplemented with 1% LNnT. An extension product was identified 59 nucleotides 5′ of the predicted translational start site for the nahS gene (Fig. S1C), while the TSS upstream of nahA was identified 74 nucleotides 5′ of the predicted nahA translational start site (Fig. S1D). The nahS upstream region contained −10 and −35 hexamers just upstream of the TSS, resembling bifidobacterial promoter sequences (35, 36), while in the case of the nahA promoter region, the TSS is preceded by a sequence that resembles a canonical −10 promoter sequence, although no associated −35 hexamer could be identified.
The nag and lnp-glt loci were each deduced to contain at least two promoters, just upstream of the nagB3 (Fig. 2e) and nagK (Fig. 2f) genes and the lnpB (Fig. 2g) and gltA (Fig. 2h) genes, respectively, based on the associated genetic layout coupled to transcription patterns of the nagR1 mutant or when UCC2003 was grown on LNB (27). The TSSs of the presumed nagB3, nagK, lnpB, and gltA promoters were determined by primer extension analysis using RNA extracted from B. breve UCC2003 grown in mMRS medium supplemented with 1% LNB. An extension product was identified 155 nucleotides 5′ of the predicted translational start site for the nagB3 gene (Fig. S1E), while the transcriptional start site of nagK was identified 35 nucleotides 5′ of the predicted translational start site (Fig. S1F). An extension product was identified 43 nucleotides 5′ of the predicted translational start site for the lnpB gene (Fig. S1G), while the transcription start site for the gltA gene was identified 44 nucleotides 5′ of the predicted translational start site (Fig. S1H). All four regions contained −10 and −35 hexamers just upstream of the TSS that resembled bifidobacterial vegetative promoter recognition sequences.
Identification of regulator-operator interactions by using electromobility shift assays and in silico analyses.
In order to establish if the LntR, NahR, and NagR1 proteins directly and specifically interact with operator sequences within the identified promoter regions of the lnt, nah, and nag-lnp-glt gene clusters, respectively, electrophoretic mobility shift assays (EMSAs) were performed. For the purpose of performing EMSAs, the lntR, nahR, and nagR1 genes were first individually cloned into the nisin-inducible vector pNZ8150 with an N-terminal His tag-encoding sequence to facilitate protein expression and purification in Lactococcus lactis NZ9000 (see Materials and Methods). As was noted previously for other regulators from bifidobacteria (37–39), LntR, NahR, and NagR1 could be obtained as purified proteins but had lost their DNA-binding activity during some stage of the purification process. Thus, instead of purified protein, crude cell extracts of (nisin-induced) L. lactis NZ9000/pNZ-lntRHis, L. lactis NZ9000/pNZ-nahRHis, and L. lactis NZ9000/pNZ-nagR1His were used to carry out the EMSAs. A crude cell extract obtained from nisin-induced L. lactis NZ9000/pNZ8150 (empty vector) incubated with the respective DNA fragments was used as a negative control. The DNA fragments used were various short amplicons representing different segments of the putative promoter regions (Fig. 2; see also Table S1 in the supplemental material).
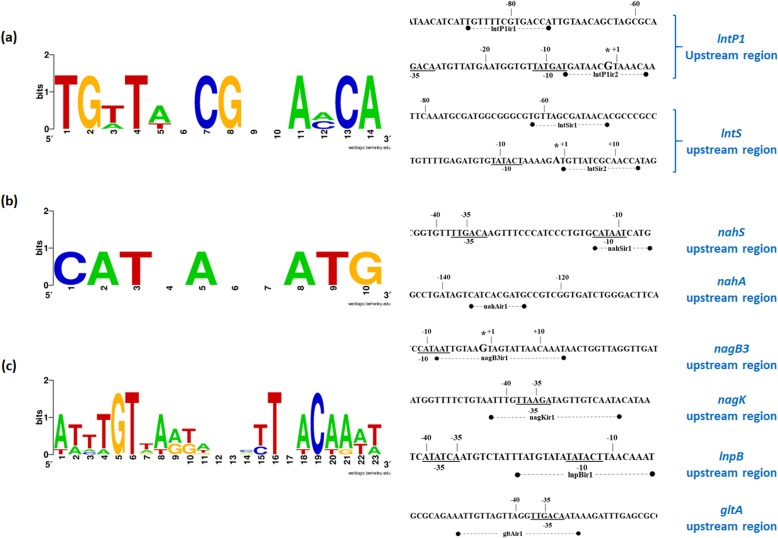
The LntR-containing crude extract was shown to specifically bind to the IRD700 (infrared dye)-labeled DNA fragments lntP1a and lntP1b but not to lntP1c (Fig. 2a and Table S1). A double mobility shift was observed for fragment lntP1a, which is indicative of two distinct LntR-binding sites being present on this fragment, while a single mobility shift was visible for fragment lntP1b. Similarly, LntR was able to bind to IRD700-labeled DNA fragments lntSb and lntSa, in the latter case being visible as a double mobility shift (suggesting the presence of two distinct LntR-binding sites), while no binding was observed with lntSc (Fig. 2b and Table S1). Inspection and comparison of the four fragments in which binding was observed revealed the presence of at least one complete conserved sequence, representing an inverted repeat, in all four fragments, while two such conserved sequences were observed in fragments lntP1a and lntSa (consistent with the observed double mobility shift). Comparative analysis of these inverted repeats identified a 14-nucleotide consensus sequence (Fig. 3a) containing conserved “CG” nucleotides at its center, which is a well-documented conserved feature of operator sequences bound by LacI-type regulators (40, 41). This consensus sequence furthermore contains conserved 5′ “TG” and 3′ “CA” nucleotides at its flanking ends, a feature previously documented for operator sequences identified for other LacI-type regulators encoded by B. breve UCC2003 (38, 42, 43). In both promoter regions, one such presumed operator sequence was found closely downstream of or partially overlapping the predicted −10 element of the promoter region, while the second was found closely upstream of the predicted −35 element of the promoter region (Fig. S1A and S1B). The positions of these identified operators are consistent with LntR acting as a repressor for the identified lnt promoters (44, 45).
FIG 3.
WebLogo representation of the operator motif consensus sequences of the LacI-type regulator LntR (a), the NahC/XylR-type regulator NahR (b), and the ROK/NagC-type regulator NagR1 (c), predicted by using in silico analysis. Predicted operator sequences were identified in the intergenic regions containing the coregulated promoters for each regulator by using the MEME online tool. Motif consensuses were generated by inputting these predicted operator sequences to the WebLogo online tool. The locations and sequences of each operator are shown alongside their respective consensus sequences.
The results obtained with the L. lactis NZ9000/pNZ-nahRHis crude extract demonstrated specific binding to the IRD700-labeled DNA fragments nahSa and nahSb but not to nahSc (Fig. 2c and Table S1). Furthermore, binding was observed for IRD700-labeled DNA fragment nahAa but not for fragment nahAb or nahAc (Fig. 2d and Table S1). Sequence inspection and comparison of the NahR-bound DNA fragments revealed the presence of an inverted-repeat sequence, which was common to these fragments yet not present in fragments to which NahR did not bind. These inverted-repeat elements therefore represent putative operator sequences required for the NahR protein. Further analysis of these inverted repeats identified a 10-nucleotide consensus sequence (Fig. 3b). Conserved 5′ C and 3′ G nucleotides at the extreme flanks of this consensus sequence were previously observed for operator sequences of certain NagC/XylR-type regulators (46). The presumed operator upstream of nahS overlaps the downstream end of the predicted −10 promoter element (Fig. S1C), while the nahA-associated operator was found to be roughly 110 bp upstream of the predicted −10 element (Fig. S1D). The position of the identified nahS operator and the consensus obtained between this and the putative operator identified for nahA confirm the function of NahR as a repressor of nahS. While binding of NahR may occur at the nahA operator, this binding does not appear to directly interfere with the nahA promoter. This agrees with the lack of upregulation of nahA expression observed for the nahR mutant, although the transcriptional role, if any, of NahR in this case is not clear. The mapped locations of the operator and −10 and −35 sequences are shown in Fig. S1 in the supplemental material.
The results obtained with the crude extract obtained from nisin-induced L. lactis NZ900/pNZ-nagR1His revealed specific binding to the IRD700-labeled DNA fragments nagB3a, nagB3b, and nagB3c (with a weak apparent double shift observed for nagB3a) but not to fragment nagB3d (Fig. 2e and Table S1). Specific binding was identified for IRD700-labeled DNA fragment nagKa, while no binding was detected when fragment nagKb was used (Fig. 2f and Table S1). Binding of the NagR protein was also demonstrated for the IRD700-labeled DNA fragments lnpBa and lnpBc but not for lnpBb (Fig. 2g and Table S1). Finally, NagR1 was shown to bind IRD700-labeled DNA fragments gltAa, gltAb, and gltAc (Fig. 2h and Table S1). Inspection and comparison of the nagB3-, nagK-, lnpB-, and gltA-associated fragments in which binding was observed revealed the presence of a common sequence, representing an inverted repeat (with two repeats present in fragment nagB3a, consistent with the observed double shift), which was absent within fragments for which no binding was observed. These sequence motifs are presumed to act as operator sequences for the NagR1 protein. In silico analysis of these inverted-repeat sequences revealed a 23-nucleotide consensus motif (Fig. 3c). Interestingly, while this obtained consensus motif bears little resemblance to many previously proposed binding motifs for ROK/NagC family-type repressors from other bacteria (47), a substantial degree of similarity to motifs identified previously for other ROK/NagC-type regulators encoded by B. breve UCC2003 can be observed (30, 38). The putative nagB3, nagK, lnpB, and gltA operators were all found to overlap or encompass the predicted −10 or −35 elements (Fig. S1E to S1H). The positions of these identified operators corroborate the notion that NagR1 acts as a transcriptional repressor of its target genes (i.e., nagB3, nagK, lnpB, and gltA).
Identification of transcriptional effectors.
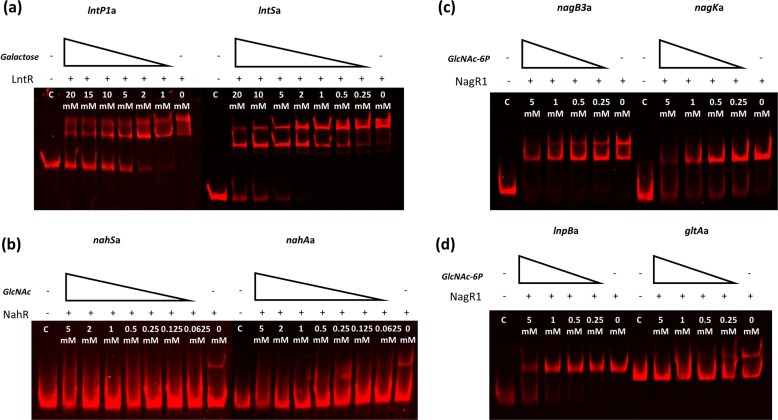
In order to identify effectors that control the binding activity of LntR, NahR, and NagR1, we performed EMSAs with fragments containing the binding motifs for each regulator, in the presence of a range of carbohydrates, including lactose, LNB, LNT, LNnT, galactose, galactose-6-phosphate (Gal-6-P), galactose-1-phosphate (Gal-1-P), GlcNAc, N-acetylglucosamine-6-phosphate (GlcNAc-6P), GalNAc, or glucose (at a standard concentration of 20 mM) (see Fig. S2 in the supplemental material). These carbohydrates were chosen as they include both the complete structures and various components (or breakdown products) of LNT, LNnT, or LNB. Carbohydrates that did not elicit any effect on fragment binding by the regulator (at a concentration of 20 mM) were assumed not to represent transcriptional effectors for that particular regulator. If inhibition of binding was observed at 20 mM, the EMSA was repeated with a range of descending concentrations (or in some cases, higher concentrations were used for a related molecule [e.g., Gal, Gal-1-P, and Gal-6-P]). For LntR, galactose was found to reduce the binding of this regulator to its DNA targets at a concentration of 10 mM or lower (Fig. 4a). Gal-6-P and Gal-1-P were also found to reduce target DNA binding of LntR but at considerably higher, and perhaps biologically irrelevant, concentrations of ≥20 mM (Fig. S3). For NahR, only GlcNAc was found to reduce the interaction between NahR and its DNA target at a minimum concentration of 0.0625 mM (Fig. 4b), while in the case of NagR1, GlcNAc-6-P was found to prevent NagR1-binding activity at a minimum concentration of 1 mM (Fig. 4c and d).
FIG 4.
EMSA images showing LntR (a), NahR (b), and NagR1 (c and d) interactions with promoter-containing DNA fragments, with the addition of a gradient of their respective inducers ranging from 0 mM to 20 mM. In each panel, “C” indicates a negative control, where an equivalent amount of the crude cell extract from NZ9000 harboring empty plasmid pNZ8150 was added instead of the crude extract from the regulator-expressing NZ9000 strain.
DISCUSSION
The dominance of (certain) bifidobacteria within the breastfed neonatal gut microbiota (1) is substantially aided by the ability of these infant-associated species to utilize indigestible HMO residues as a carbon source (9). Our previous work demonstrated that the consumption and utilization of LNT, LNnT, or LNB by B. breve UCC2003 are facilitated by interrelated catabolic pathways (27). While pathways for HMO utilization in other Bifidobacterium species have been identified and elucidated (13, 21, 22), very little work has been carried out with regard to their regulation. Our results reveal molecular details of the transcriptional regulation of B. breve UCC2003 loci responsible for LN(n)T/LNB metabolism and provide insights into how the metabolism of these HMOs is controlled in B. breve UCC2003.
In this study, we identified four transcriptional regulators, three of which were shown to be involved in regulating LN(n)T/LNB metabolism in UCC2003 (Fig. 5). Microarray analysis of insertional mutants of lntR, nahR, nagR1, and nagR2 identified genes under the regulation of each encoded regulator. LntR and NahR were shown to represent “local” regulators, i.e., controlling the transcription of genes adjacent to lntR and nahR, respectively. In contrast, NagR1 regulates the transcription of not only the local nag locus but also the genetically unlinked lnp-glt locus. We also investigated the transcriptome effect of a mutation in Bbr_1251 (nagR2); however, the affected genes are not believed to be involved in HMO metabolism but apparently are involved in maltooligosaccharide and fatty acid metabolism. While LacI-type, NagC/XylR-type, and ROK/NagC-type regulators have all previously been identified and characterized in B. breve UCC2003 (30, 37, 38, 42), functional analysis of regulators in other bifidobacteria is comparatively undocumented. However, a recent study identified transcription factors homologous to those of LntR, NagR1, and NagR2 in a range of different Bifidobacterium species (48).
FIG 5.
Schematic representation of the proposed model for the transcriptomic regulation of LNT, LNnT, and LNB metabolism by B. breve UCC2003. LNT and LNnT are internalized and intracellularly degraded through the sequential hydrolysis and release of monosaccharides from their nonreducing ends. These released monosaccharides (or their metabolites) act as effectors to relieve the transcriptional repression of the loci encoding the cellular components responsible for the liberation of these glycans. As such, liberated galactose relieves the transcriptional repression of the lnt locus, and N-acetylglucosamine relieves the transcriptional repression of the nah locus. Similarly, the intracellular degradation of LNB (derived from the extracellular hydrolysis of complex HMO structures by other infant gastrointestinal tract microbes) releases N-acetylglucosamine and galactose-1-phosphate. GlcNAc is further converted into GlcNAc-6-P, which relieves the transcriptional repression of the lnp-glt and nag loci.
Details of promoter and operator sequences specific to the LntR, NahR, and NagR1 regulators were elucidated by using a combination of electromobility shift and primer extension analyses. These operator results, for the most part, agree with those predicted previously by Khoroshkin et al. (48). The operator sequences predicted in their study concur with our experimentally determined data, in both approximate location and number, for both LntR and NagR1, with the exception of one additional predicted operator for LntR and two for NagR1. An additional NagR1 operator sequence was predicted to be upstream of gltA; however, this did not appear to be functional, based on the lack of a double mobility shift in the EMSAs of this region. This operator may indeed be a nonfunctional relic resulting from a duplication event. Khoroshkin et al. (48) also predicted an operator sequence upstream of the Bbr_1884 gene for NagR1 binding, although we did not examine this. However, based on the predicted functions of this gene in the bifid shunt, it may also be tied into the overall regulation of LNB and LacNAc metabolism carried out by NagR1. An additional LntR operator was predicted to be upstream of lntR itself, which may function in lntR transcriptional autoregulation. The observed lack of upregulation of nahA transcription for the nahR mutant appears discordant with the increase in the transcriptional level of this gene that was previously observed for wild-type UCC2003 during growth on LN(n)T (27) as well as the presence of the functional nahA operator sequence for NahR binding identified in this study. However, this may be explained if the transcriptional induction of nahA is mediated by both LntR and NahR. This possibility is corroborated by the presence of an inverted-repeat sequence resembling an LntR operator, and this intriguing possibility merits further experimental investigation.
Perhaps most interesting of all was the identification of the effectors for each transcriptional regulator. The binding of LntR to its targets is impeded by Gal, and NahR-mediated operator binding is prevented by the presence of GlcNAc, while the NagR1-operator interaction is prevented by the presence of GlcNAc-6-P. In each case, the genes under the transcriptional control of their respective regulators encode the metabolic machinery responsible for the release (and/or generation) of the effector monosaccharide from the substrate at that metabolic step. For example, Gal is released from the nonreducing end of LN(n)T through the hydrolytic activity of LntA, which is encoded by the lnt locus (27). The transcriptional repression of this locus is thus believed to be relieved by the presence of the released monosaccharide, which is presumed to interact with the allosteric effector site typical of LacI-type repressors (38, 44, 49, 50). A similar scenario applies to GlcNAc release, which acts as the effector for the NahR regulator that controls the transcription of nahS, and to GlcNAc-6-P, which governs the activity of NagR1, the presumed transcriptional regulator of the lnp-glt and nag loci. The possible dual regulation of nahA transcription, as mentioned above, would mean that the presence of both the lnt locus activity product (and LntR effector) galactose and the nah locus activity product (and NahR effector) GlcNAc is required for the induction of nahA expression. This provides an extra level of transcriptional and, thus, metabolic control, ensuring the expression of nahA strictly during LN(n)T metabolism despite GlcNAc release during the metabolism of other sugars, such as LNB, sialic acid, and sulfated GlcNAc (27, 29, 30). Interestingly, in the case of GlcNAc-6-P and NagR1, the lnp-glt locus is required for the degradation of LNB, while the activity of the nag locus results in the generation of GlcNAc-6-P from liberated GlcNAc, during metabolism of both HMO and sialic acid (29). This may not be surprising, as sialic acid residues are commonly found in HMOs (6), and more importantly, GlcNAc is a breakdown product of LNB (as well as LNT and LNnT).
Interestingly, previous work showed that transcriptional induction takes place at the lnt locus during the growth of UCC2003 on galacto-oligosaccharides (GOSs) (51). This would appear to disagree with the high degree of specificity of transcriptional induction by effectors of these HMO-associated loci. However, it is worth noting that GOSs consist mainly of galactose (52, 53) and that the intracellular release of galactose during GOS metabolism by UCC2003 would be sufficient to cause the transcriptional induction of the lnt locus.
Thus, the presence and initial degradation of such a structure (i.e., LNT, LNnT, or LNB) indirectly induce the further expression of the locus required for its degradation, until the sugar is no longer available, at which point the absence of inducers will cause a return to transcriptional repression. Initial internalization and degradation are likely facilitated by a low level of “leaky” gene expression of the locus. In the case of LNT and LNnT degradation, this regulation is a two-step process, first at the level of LN(n)T degradation (by the lnt locus) and then at the level of [LN(n)T breakdown product] lacto-N-triose degradation (by the nah locus). The regulation of LNB metabolism is managed in a single step, at the level of LNB phosphorolysis and GlcNAc phosphorylation (by the lnp-glt locus and the nag locus, respectively). We see that all three regulators in this transcriptional control network belong to distinct families of regulator proteins despite functioning in similar roles as saccharide-controlled repressors. In conclusion, our results reveal a tightly controlled system for the transcriptional regulation of genes encoding the metabolic machinery required for (certain) HMO metabolism in B. breve UCC2003. Such tight regulation is necessary for infant-associated bifidobacteria such as B. breve, where switching metabolic processing to and from milk-derived sugars, such as HMO and lactose, and plant-derived carbohydrate sources (54) is a regular occurrence during the weaning period. Moreover, this suggests the evolution of specific catabolic responses to the presence of and for the utilization of specific HMO moieties by B. breve and poses the question of whether such regulatory systems have similarly evolved in other infant-associated Bifidobacterium species.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
Bacterial strains and plasmids used in this study are listed in Table 2. B. breve UCC2003 was routinely cultured in either de Man-Rogosa-Sharpe (MRS) medium (Difco, BD, Le Pont de Claix, France) supplemented with 0.05% cysteine-HCl or reinforced clostridial medium (RCM; Oxoid Ltd., Basingstoke, England). Growth of bifidobacterial strains for transcriptional and primer extension analyses was carried out with mMRS medium, which was prepared from first principles (using individual components) (55) and which does not contain a fixed carbohydrate source. Prior to inoculation, mMRS medium was supplemented with cysteine-HCl (0.05%, wt/vol) and a particular carbohydrate source (1%, wt/vol). It was previously shown that mMRS medium does not support the growth of B. breve UCC2003 in the absence of an added carbohydrate (56). Carbohydrates used were ribose (Sigma-Aldrich, Steinheim, Germany), LNB (Elicityl Oligotech, Crolles, France), and LNnT (Glycom, Lyngby, Denmark). A 1% (wt/vol) concentration of the carbohydrate was considered sufficient to encourage adequate growth for RNA harvesting. The addition of these carbohydrates did not significantly alter the pH of the medium, and therefore, subsequent pH adjustment was not required.
TABLE 2.
Bacterial plasmids and strains used in this worka
| Strain or plasmid | Relevant feature(s) | Reference |
|---|---|---|
| Strains | ||
| Escherichia coli | ||
| EC101 | Cloning host; repA+ Km+ | 42 |
| EC101/pNZ-M.BbrII+M.BbrIII | EC101 harboring pNZ8048 derivative containing bbrIIM and bbrIIIM | 40 |
| Lactococcus lactis | ||
| NZ9000 | MG1363 pepN::nisRK; nisin-inducible overexpression host | 54 |
| NZ9700 | Nisin-producing strain | 54 |
| NZ9000/pNZ-lntR | NZ9000 containing pNZ-lntR | This study |
| NZ9000/pNZ-nahR | NZ9000 containing pNZ-nahR | This study |
| NZ9000/pNZ-nagR1 | NZ9000 containing pNZ-nagR1 | This study |
| Bifidobacterium breve | ||
| UCC2003 | Isolate from nursling stool | 41 |
| UCC2003-lntR | pORI19-tet-bbr_0526 insertion mutant of UCC2003 | This study |
| UCC2003-nahR | pORI19-tet-bbr_1555 insertion mutant of UCC2003 | This study |
| UCC2003-nagR1 | pORI19-tet-bbr_1249 insertion mutant of UCC2003 | This study |
| UCC2003-nagR2 | pORI19-tet-bbr_1251 insertion mutant of UCC2003 | This study |
| Plasmids | ||
| pAM5 | pBC1-puC19-Tetr | 82 |
| pORI19 | Emr ΔrepA ori+; cloning vector | 42 |
| pORI19-tet-lntR | Internal 367-bp fragment of Bbr_0526 and tetW cloned into pORI19 | This study |
| pORI19-tet-nahR | Internal 448-bp fragment of Bbr_1554 and tetW cloned into pORI19 | This study |
| pORI19-tet-nagR1 | Internal 502-bp fragment of Bbr_1249 and tetW cloned into pORI19 | This study |
| pORI19-tet-nagR2 | Internal 507-bp fragment of Bbr_1251 and tetW cloned in pORI19 | This study |
| pNZ8150 | Cmr; nisin-inducible translational fusion vector | 50 |
| pNZ-lntR | Cmr; pNZ8150 derivative containing a translational fusion of the Bbr_0526 DNA fragment to the nisin-inducible promoter | This study |
| pNZ-nahR | Cmr; pNZ8150 derivative containing a translational fusion of the Bbr_1555 DNA fragment to the nisin-inducible promoter | This study |
| pNZ-nagR1 | Cmr; pNZ8150 derivative containing a translational fusion of the Bbr_1249 DNA fragment to the nisin-inducible promoter | This study |
Cmr, Emr, Kmr, and Tetr indicate resistance to chloramphenicol, erythromycin, kanamycin, and tetracycline, respectively. UCC, University College Cork Culture Collection.
B. breve cultures were incubated under anaerobic conditions in a modular atmosphere-controlled system (Davidson and Hardy, Belfast, Ireland) at 37°C. Lactococcus lactis strains were cultivated in M17 broth (Oxoid Ltd., Basingstoke, England) containing 0.5% glucose (57) at 30°C. Escherichia coli strains were cultured in Luria-Bertani (LB) broth (58) at 37°C with agitation. Where appropriate, growth media contained tetracycline (Tet) (10 μg ml−1), chloramphenicol (Cm) (5 μg ml−1 for L. lactis and E. coli and 2.5 μg ml−1 for B. breve), erythromycin (Em) (100 μg ml−1), or kanamycin (Kan) (50 μg ml−1). Recombinant E. coli EC101 cells containing (derivatives of) pORI19 were selected on LB agar containing Em and Kan and supplemented with X-gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) (40 μg ml−1) and 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside).
Nucleotide sequence analysis.
Sequence information was obtained from the Artemis-mediated (59) genome annotations of B. breve UCC2003 (60). Database searches were performed by using nonredundant sequences accessible at the National Center for Biotechnology Information (NCBI) (http://www.ncbi.nlm.nih.gov/), using the basic local alignment search tool (BLAST) (61, 62). Sequences were verified and analyzed by using the SeqMan and SeqBuilder programs of the DNAStar software package (version 10.1.2; DNAStar, Madison, WI, USA).
DNA manipulations.
Chromosomal DNA was isolated from B. breve UCC2003 as previously described (63). Plasmid DNA was isolated from Escherichia coli, Lactococcus lactis, and B. breve by using the Roche High Pure plasmid isolation kit (Roche Diagnostics, Basel, Switzerland). An initial lysis step was performed by using 30 mg ml−1 of lysozyme for 30 min at 37°C prior to plasmid isolation from L. lactis or B. breve. Procedures for DNA manipulations were performed essentially as described previously (58). All restriction enzymes and T4 DNA ligase were used according to the supplier's instructions (Roche Diagnostics, Basel, Switzerland). Synthetic single-stranded oligonucleotide primers used in this study (Table 3) were synthesized by Eurofins (Ebersberg, Germany). Standard PCRs were performed by using Taq PCR master mix (Qiagen) or Extensor Hi-Fidelity PCR master mix (Thermo Scientific, Waltham, MA, USA) in a Biometra T3000 thermocycler (Biometra, Göttingen, Germany) or a Life Technologies Proflex PCR system (Thermo Scientific, Waltham, MA, USA). PCR products were visualized by ethidium bromide (EtBr) staining following agarose gel electrophoresis (1% agarose). B. breve colony PCRs were performed as described previously (64). PCR fragments were purified by using the Roche High Pure PCR purification kit (Roche Diagnostics, Basel, Switzerland). Plasmid DNA was isolated by using the Roche High Pure plasmid isolation kit (Roche Diagnostics, Basel, Switzerland). Plasmid DNA was introduced into E. coli by electroporation, as described previously (58). B. breve UCC2003 (65) and L. lactis (66) were transformed by electroporation according to previously reported protocols. The correct orientation of DNA inserts and the integrity of all plasmid constructs (see also below) were verified by DNA sequencing performed at Eurofins (Ebersberg, Germany).
TABLE 3.
Oligonucleotide primers used in this work
| Purpose | Primer | Sequence (5′–3′)a |
|---|---|---|
| Cloning of Bbr_0526 in pNZ8150 | 526F | TGCATCCCCGGGATGCATCACCATCACCATCACCATCACCATCACGCGAGACCAACACAGGTTTCC |
| 526R | TGCGCATCTAGACGTTTCCCGTATACCATTAATCAG | |
| Cloning of Bbr_1555 in pNZ8150 | 1555F | TGCATCGATATCATGCATCACCATCACCATCACCATCACCATCACTACGCTAAATCCAATCCC |
| 1555R | TGCGCATCTAGACGGCGGCACGGTGATCTG | |
| Cloning of Bbr_1249 in pNZ8150 | 1249F | TGCATCCAGCTGATGCATCACCATCACCATCACCATCACCATCACTCGTATCCCGGTCTTGCC |
| 1249R | TGCATCCAGCTGATGTCGTATCCCGGTCTTGCC | |
| Cloning of an internal 465-bp fragment of Bbr_0526 in pORI19 | IM526F | CTGGTCAAGCTTCGTTGAAGCCGCGATGGA |
| IM526R | CTGGTCTCTAGAGTCAACGGTGGGGCAGTG | |
| Cloning of an internal 488-bp fragment of Bbr_1555 in pORI19 | IM1555F | CTGGTCAAGCTTGCTGGCCATCGATACGGAC |
| IM1555R | CTGGTCTCTAGACTCGTCGTTCAGCAGCAC | |
| Cloning of an internal 443-bp fragment of Bbr_1249 in pORI19 | IM1249F | CTGGTCAAGCTTCGAAGAAGGCCTATTGCG |
| IM1249R | CTGGTCTCTAGACAGCAGAATCGCCGAACC | |
| Cloning of an internal 488-bp fragment of Bbr_1251 in pORI19 | IM1251F | CTGGTCAAGCTTGAAGAGACCGGCGACCTGG |
| IM1251R | CTGGTCTCTAGAGCCATTGTCGATGACGCC | |
| Amplification of tetW | tetWFw | TCAGCTGTCGACATGCTCATGTACGGTAAGGAAGCA |
| tetWRv | GCGACGGTCGACCATAACTTCTGATTGTTGCCG | |
| Confirmation of site-specific homologous recombination | 526confirm1 | GCGCTAGCTGTTACAATGGTC |
| 526confirm2 | GCCATTTCCAACCCCTCTC | |
| 1555confirm1 | TACGCTAAATCCAATCCC | |
| 1555confirm2 | GACGCAAGGGCCAACAACCGC | |
| 1249confrim1 | CATACAGCCGCCACGGCAC | |
| 1249confrim2 | TCGTATCCCGGTCTTGCC | |
| 1251confrim1 | GCAGACGATACTGCACGCG | |
| 1251confrim2 | GTCAAGCATCTCTACCAC | |
| Amplification of Bbr_0527 promoter fragments with IRD700-labeled oligonucleotides | 527IRDfa | CTCGCCCCTCGCTTGTCTCTC |
| 527IRDra | GCATAGGCACGGCAGCGAC | |
| 527IRDfb | ATTGTTTTCGTGACCATTG | |
| 527IRDrb | GAATAATGAACACGAACACG | |
| 527IRDfc | CAATTTTGGTCAACCTTCG | |
| 527IRDrc | CGCGCGTAGTTCTCGAC | |
| Amplification of Bbr_0530 promoter fragments with IRD700-labeled oligonucleotides | 530IRDfa | GCCGAACGGTGTGCTGGTGG |
| 530IRDra | CTTCATCGTTCTGTTCTCCTTC | |
| 530IRDfb | CGATAACACGCCCGCCATC | |
| 530IRDrb | GCTGGACTTGCCGCTATC | |
| 530IRDfc | CTTCATAGAGCCACTTC | |
| 530IRDrc | CTCGAAGTCCTTGGCAAC | |
| Amplification of Bbr_1554 promoter fragments with IRD700-labeled oligonucleotides | 1554IRDfa | GTCGCTGGGATTGGATTTAGCG |
| 1554IRDra | GTGGCTATGACTGCGCGC | |
| 1554IRDfb | CGGCTTTCAGGATAACACCCA | |
| 1554IRDrb | GGATTTGGCGGCGCGATC | |
| 1554IRDfc | CCAAACAAAATAGTTGCTACGGC | |
| 1554IRDrc | GTTGAGTGCGGTGTAGGTCTCC | |
| Amplification of Bbr_1556 promoter fragments with IRD700-labeled oligonucleotides | 1556IRDfa | CTGGACGGCTGCTCAAAGC |
| 1556IRDra | GCAGAGATGTTTGACCGTTCAT | |
| 1556IRDfb | GGTGACGACGCCACTCTGC | |
| 1556IRDrb | GTGGTTTGCGGTTGCCCT | |
| 1556IRDfc | GCCATCTCAGGACCGAACG | |
| 1556IRDrc | GGTCGTCAAGGTGATGAATCC | |
| Amplification of Bbr_1248 promoter fragments with IRD700-labeled oligonucleotides | 1248IRDfa | CCTCCTGCCTGAACGATG |
| 1248IRDra | GACAATGATGATTTCCGGC | |
| 1248IRDfb | GTTAGGGAACTTCACTAATACATTCC | |
| 1248IRDrb | CCTGGGAGATGTCGATCGACTC | |
| 1248IRDfc | GTCCGTACGTCCATAATTGTAAGTAG | |
| 1248IRDrc | CCTGGGAGATGTCGATCGACTC | |
| 1248IRDfd | GATGGGCGGCTTTGGGCAG | |
| 1248IRDrd | CCTGGGAGATGTCGATCGACTC | |
| Amplification of Bbr_1250 promoter fragments with IRD700-labeled oligonucleotides | 1250IRDfa | GATGCCGTTGTGGTAGAGATG |
| 1250IRDra | GGTGTTATCAGTCATTGCCTATCC | |
| 1250IRDfb | GCGTGTCGCGTATGAGGC | |
| 1250IRDrb | GGTGTTATCAGTCATTGCCTATCC | |
| Amplification of Bbr_1586 promoter fragments with IRD700-labeled oligonucleotides | 1586IRDfa | CGGTTCGTCGAAAATCCAAG |
| 1586IRDra | CAGTGCGAAGTGTGAGGCG | |
| 1586IRDfb | GCCGCTTATTGCGGCTTTATAG | |
| 1586IRDrb | CTTTGAGGGCAGAAGTAACTAGTTC | |
| 1586IRDfc | GTATGCGCGTTCGTCCAC | |
| 1586IRDrc | CTTTGAGGGCAGAAGTAACTAGTTC | |
| Amplification of Bbr_1590 promoter fragments with IRD700-labeled oligonucleotides | 1590IRDfa | GGCCCGCTGGCAGATTAG |
| 1590IRDra | GGCAAGAGCAGCCACGATG | |
| 1590IRDfb | GACAGATGTCTGAGCGGTC | |
| 1590IRDrb | GAATCGGGCAGACGGTGC | |
| 1590IRDfc | CGCGCAGAAATTGTTAGTTAGG | |
| 1590IRDrc | GAATCGGGCAGACGGTGC | |
| Amplification of a region containing the Bbr_0527 promoter region for sequencing ladders | 527promF | GCATTGCTGTCATTCGCCACAC |
| 527promR | GAATAATGAACACGAACACG | |
| Amplification of a region containing the Bbr_0530 promoter region for sequencing ladders | 530promF | GCGTGCGGATGAAACTGG |
| 530promR | GTCTGGAACGGCTTGGCGC | |
| Amplification of a region containing the Bbr_1554 promoter region for sequencing ladders | 1554promF | CGTTTCCTCGACCCCAGTTC |
| 1554promR | GAATGTGTCCTTGAGCTTGGC | |
| Amplification of a region containing the Bbr_1556 promoter region for sequencing ladders | 1556promF | CTGGACGGCTGCTCAAAGC |
| 1556promR | GGTCGTCAAGGTGATGAATCC | |
| Amplification of a region containing the Bbr_1248 promoter region for sequencing ladders | 1248promF | GGAGGCTTTGGCGGTACGG |
| 1248promR | CCTGGGAGATGTCGATCGACTC | |
| Amplification of a region containing the Bbr_1250 promoter region for sequencing ladders | 1250promF | GATGCCGTTGTGGTAGAGATG |
| 1250promR | GGTGCCACCCACATCAACAC | |
| Amplification of a region containing the Bbr_1586 promoter region for sequencing ladders | 1586promF | GCGAGACCTTCGACCTTCAGCC |
| 1586promR | CGGCACGAGATTGTAAGACAC | |
| Amplification of a region containing the Bbr_1590 promoter region for sequencing ladders | 1590promF | GGCCCGCTGGCAGATTAG |
| 1590promR | GAATCGGGCAGACGGTGC | |
| 527 promoter for primer extension analysis | 527PE | GCATAGGCACGGCAGCGAC |
| 530 promoter for primer extension analysis | 530PE | CTTCATCGTTCTGTTCTCCTTC |
| 1554 promoter for primer extension analysis | 1554PE | GTTCATGTTGGTCTTCTTTCC |
| 1556 promoter for primer extension analysis | 1556PE | GCAGAGATGTTTGACCGTTCAT |
| 1248 promoter for primer extension analysis | 1248PE | CTGCCCAAAGCCGCCCATC |
| 1250 promoter for primer extension analysis | 1250PE | GGTGTTATCAGTCATTGCCTATCC |
| 1586 promoter for primer extension analysis | 1586PE | GCGATGTCAAATAGTGTTTCC |
| 1590 promoter for primer extension analysis | 1590PE | GGCAAGAGCAGCCACGATG |
Restriction sites incorporated into oligonucleotide primer sequences are indicated in boldface type, and His-tagged sequences incorporated into nucleotide primer sequences are indicated in italic type.
Construction of B. breve UCC2003 insertion mutants.
Internal fragments of Bbr_0526 (designated here lntR) (367 bp representing codons 40 through 162 of the 320 codons of this gene), Bbr_1249 (designated here nagR1) (502 bp representing codons 64 through 231 of the 375 codons of this gene), Bbr_1251 (designated here nagR2) (507 bp representing codons 62 through 230 of the 405 codons of this gene), and Bbr_1555 (designated here nahR) (448 bp representing codons 74 through 223 of the 380 codons of this gene) were amplified by PCR using B. breve UCC2003 chromosomal DNA as a template and primer pairs 526LacIInsFHindIII and 526LacIInsRXbaI, 1249LacIInsFHindIII and 1249LacIInsRXbaI, 1251LacIInsFHindIII and 1251LacIInsRXbaI, and 1555LacIInsFHindIII and 1555LacIInsRXbaI (Table 3), respectively. The insertion mutants were constructed by using a previously described approach (64), generating mutant strains B. breve UCC2003-lntR, B. breve UCC2003-nagR1, B. breve UCC2003-nagR2, and B. breve UCC2003-nahR, which carried disrupted lntR, nagR1, nagR2, and nahR genes, respectively (Table 2). The site-specific recombination of potential Tet-resistant mutant isolates was confirmed by colony PCR using primer pair TetWF and TetWR to verify tetW gene integration and primers Bbr_526ConfirmP1 or Bbr_526ConfirmP2, Bbr_1249ConfirmP1 or Bbr_1249ConfirmP2, Bbr_1251ConfirmP1 or Bbr_1251ConfirmP2, and Bbr_1555ConfirmP1 or Bbr_1555ConfirmP2 (positioned upstream of the selected internal fragments of Bbr_0526, Bbr_1249, Bbr_1251, and Bbr_1555, respectively) in combination with primer TetWF to confirm integration at the correct chromosomal location (Table 3).
Analysis of global gene expression using B. breve DNA microarrays.
Global gene expression levels were determined during log-phase growth of the insertional mutant strains B. breve UCC2003-lntR, B. breve UCC2003-nagR1, B. breve UCC2003-nagR2, and B. breve UCC2003-nahR in mMRS medium supplemented with ribose. The generated transcriptome data sets were compared to the transcriptome information obtained for log-phase wild-type B. breve UCC2003 cells grown in mMRS medium supplemented with ribose. Ribose was selected as a suitable transcriptomic reference as the metabolic pathway and gene expression profile for the growth of UCC2003 on ribose are known and were employed previously (43, 67). DNA microarrays containing oligonucleotide primers representing each of the 1,864 identified open reading frames on the genome of B. breve UCC2003 were designed by and obtained from Agilent Technologies (Palo Alto, CA, USA). Methods for cell disruption, RNA isolation, RNA quality control, and cDNA synthesis and labeling were performed as described previously (68). Two independent biological replicates were used for each array, using a Cy3/Cy5 dye swap, as described previously (68). Labeled cDNA was hybridized by using the Agilent gene expression hybridization kit (part no. 5188-5242), as described in the Agilent Two-Color Microarray-Based Gene Expression Analysis v4.0 manual (publication number G4140-90050). Following hybridization, microarrays were washed in accordance with Agilent's standard procedures and scanned by using an Agilent DNA microarray scanner (model G2565A). The generated scans were converted to data files with Agilent Feature Extraction software (version 9.5). DNA microarray data were processed as previously described (69–71). Differential expression tests were performed with the Cyber-T implementation of a variant of the t test (72).
Construction of overexpression vectors and protein overproduction and purification.
For the construction of plasmids pNZ-lntR, pNZ-nagR1, and pNZ-nahR, DNA fragments encompassing lntR, nagR1, and nahR were generated by PCR amplification from chromosomal DNA of B. breve UCC2003 using Q5 High-Fidelity DNA polymerase and primer pairs 526PurFSmaI and 526PurRXbaI, 1249PurFPvuII and 1249PurRXbaI, and 1555PurFEcoRV and 1555PurXbaI, respectively (Table 3). An in-frame N-terminal His10-encoding sequence was incorporated into forward primers 526PurFSmaI, 1249PurFPvuII, and 1555PurFEcoRV to facilitate downstream protein purification. The generated amplicons were digested with SmaI and XbaI, PvuII and XbaI, and EcoRV and XbaI, respectively, and ligated into the ScaI- and XbaI-digested, nisin-inducible translational fusion plasmid pNZ8150 (73). The ligation mixtures were introduced into L. lactis NZ9000 by electrotransformation, and transformants were then selected based on Cm resistance. The plasmid content of a number of Cm-resistant transformants was screened by restriction analysis, and the integrity of positively identified clones was verified by sequencing.
Nisin-inducible gene expression and protein overproduction were performed as described previously (37, 42, 74). In brief, 50 ml of M17 broth supplemented with 0.5% (wt/vol) glucose was inoculated with a 2% inoculum of a particular L. lactis strain, followed by incubation at 30°C until an optical density at 600 nm (OD600) of 0.5 was reached, at which point protein expression was induced by the addition of the cell-free supernatant of a nisin-producing strain (75), followed by continued incubation for a further 2 h. Cells were harvested by centrifugation, and the crude cell extract was obtained as described previously (38). Although protein purification of LntR-His, NahR-His, and NagR1-His was achieved by using His tag affinity chromatography, the purification procedure appeared to render the proteins inactive in subsequent EMSAs. For this reason, crude cell extracts, prepared in 10 mM Tris-HCl lysis buffer (pH 7.0), were adopted for the EMSAs (see below).
Electrophoretic mobility shift assays.
DNA fragments representing different portions of the promoter regions upstream of lntP1 (locus tag Bbr_0527) and lntS (locus tag Bbr_0530), nagB3 (locus tag Bbr_1248) and nagK (locus tag Bbr_1250), lnpB (locus tag Bbr_1586) and gltA (locus tag Bbr_1590), and nahS (Bbr_1554) and nahA (Bbr_1556) were prepared by PCR using IRD700-labeled primer pairs (Integrated DNA Technologies, Coralville, IN, USA) (Table 3). EMSAs were performed essentially as described previously (42, 76). In all cases, binding reactions were carried out with a final volume of 20 μl in the presence of poly(dI-dC) in binding buffer (20 mM Tris-HCl, 5 mM MgCl2, 0.5 mM dithiothreitol [DTT], 1 mM EDTA, 100 mM KCl, 10% glycerol). Various amounts of the crude protein extract, ranging from 140 ng to 180 ng, of the constructed LntR-, NahR-, or NagR1-(over)producing L. lactis NZ9000 strain and a fixed amount of a DNA probe (0.1 pmol) were mixed on ice and subsequently incubated for 15 min at 37°C. In order to assess if the binding activity of LntR, NahR, or NagR1 is modulated by a carbohydrate ligand, various carbohydrates, including galactose, galactose-1-phosphate, galactose-6-phosphate (all from Sigma-Aldrich, Steinheim, Germany), LNT (Glycom, Lyngby, Denmark), LNnT (Glycom, Lyngby, Denmark), LNB (Elicityl Oligotech, Crolles, France), glucose, N-acetylglucosamine, N-acetylglucosamine-6-phosphate, or lactose (all from Sigma-Aldrich, Steinheim, Germany), with concentrations ranging from 50 to 0.0625 mM, were included in the binding reaction buffer. Samples were loaded onto a 6% nondenaturing phosphonoacetic acid (PAA) gel prepared in TAE buffer (40 mM Tris acetate [pH 8.0], 2 mM EDTA) and run in a 0.5×-to-2.0× gradient of TAE at 100 V for 90 min in an Atto Mini PAGE system (Atto Bioscience and Biotechnology, Tokyo, Japan). Signals were detected by using the Odyssey infrared imaging system (Li-Cor Biosciences UK Ltd., Cambridge, UK) and captured by using the supplied Odyssey V3.0 software.
Primer extension analysis.
Total RNA was isolated from B. breve UCC2003 cells grown in mMRS medium supplemented with 1% LNnT or 1% LNB to early exponential phase, using a previously described Macaloid method (77). RNA samples were treated with RNase-free DNase (Ambion). Primer extension was performed by annealing 1 pmol of IRD700 synthetic 18-mer oligonucleotides to 15 μg of RNA, as described previously (78). Sequence ladders of the presumed promoter regions immediately upstream of lntP1, lntS, nagB3, nagK, lnpB, gltA, nahS, or nahA, amplified from UCC2003 genomic DNA, which were run alongside the primer extension products, were produced by using the same primer as the one used for the primer extension reaction and by employing the Thermo Sequenase primer cycle sequencing kit (Amersham). Separation was achieved on a 6.5% Li-Cor Matrix KB Plus acrylamide gel. Signal detection and image capture were performed by means of a Li-Cor sequencing instrument (Li-Cor Biosciences).
Operator consensus sequence prediction using MEME and WebLogo online software tools.
Coregulated promoter regions were assessed for the presence of operator sequences by the use of the MEME (Multiple Em for Motif Elicitation) online tool (http://meme-suite.org/tools/meme) (79) and then visualized by using the WebLogo online tool (http://weblogo.berkeley.edu/logo.cgi) (80, 81). Sequences used for consensus sequence prediction are given in Table S2 in the supplemental material.
Accession number(s).
The microarray data obtained in this study have been deposited in the NCBI Gene Expression Omnibus (GEO) database and are accessible through GEO series accession no. GSE105108.
Supplementary Material
ACKNOWLEDGMENTS
We sincerely thank Glycom A/S (Lyngby, Denmark) for the provision of purified HMO samples used in this study under their donation program. We thank Takane Katayama for indicating an error within the manuscript prior to its official publication.
This study was funded in part by the Irish Research Council under the Postgraduate Research Project Award (project GOIPG/2013/651). In addition, we are supported by Science Foundation Ireland (SFI) (grant no. SFI/12/RC/2273). M.O.M. is a recipient of an HRB postdoctoral fellowship (grant no. PDTM/20011/9).
D.V.S., K.J., and M.O.M. conceived the experiments. K.J., with the assistance of C.P. and R.L.O., conducted the experiments. All authors analyzed the results and contributed to writing the manuscript.
We declare that, to the best of our knowledge, there are no competing financial interests.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.02774-17.
REFERENCES
- 1.Turroni F, Peano C, Pass DA, Foroni E, Severgnini M, Claesson MJ, Kerr C, Hourihane J, Murray D, Fuligni F, Gueimonde M, Margolles A, De Bellis G, O'Toole PW, van Sinderen D, Marchesi JR, Ventura M. 2012. Diversity of bifidobacteria within the infant gut microbiota. PLoS One 7:e36957. doi: 10.1371/journal.pone.0036957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sivan A, Corrales L, Hubert N, Williams JB, Aquino-Michaels K, Earley ZM, Benyamin FW, Lei YM, Jabri B, Alegre M-L, Chang EB, Gajewski TF. 2015. Commensal Bifidobacterium promotes antitumor immunity and facilitates anti-PD-L1 efficacy. Science 350:1084–1089. doi: 10.1126/science.aac4255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Maslowski KM, Mackay CR. 2011. Diet, gut microbiota and immune responses. Nat Immunol 12:5–9. doi: 10.1038/ni0111-5. [DOI] [PubMed] [Google Scholar]
- 4.Milani C, Duranti S, Bottacini F, Casey E, Turroni F, Mahony J, Belzer C, Delgado Palacio S, Arboleya Montes S, Mancabelli L, Lugli GA, Rodriguez JM, Bode L, de Vos W, Gueimonde M, Margolles A, van Sinderen D, Ventura M. 2017. The first microbial colonizers of the human gut: composition, activities, and health implications of the infant gut microbiota. Microbiol Mol Biol Rev 81:e00036-. doi: 10.1128/MMBR.00036-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gibson GR, Roberfroid MB. 1995. Dietary modulation of the human colonic microbiota: introducing the concept of prebiotics. J Nutr 125:1401–1412. [DOI] [PubMed] [Google Scholar]
- 6.Kunz C, Rudloff S, Baier W, Klein N, Strobel S. 2000. Oligosaccharides in human milk: structural, functional, and metabolic aspects. Annu Rev Nutr 20:699–722. doi: 10.1146/annurev.nutr.20.1.699. [DOI] [PubMed] [Google Scholar]
- 7.Brüssow H. 2015. Human microbiota. ‘The philosophers have only interpreted the world in various ways. The point, however, is to change it’. Microb Biotechnol 8:11–12. doi: 10.1111/1751-7915.12259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Asakuma S, Hatakeyama E, Urashima T, Yoshida E, Katayama T, Yamamoto K, Kumagai H, Ashida H, Hirose J, Kitaoka M. 2011. Physiology of consumption of human milk oligosaccharides by infant gut-associated bifidobacteria. J Biol Chem 286:34583–34592. doi: 10.1074/jbc.M111.248138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sela DA, Mills DA. 2010. Nursing our microbiota: molecular linkages between bifidobacteria and milk oligosaccharides. Trends Microbiol 18:298–307. doi: 10.1016/j.tim.2010.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kitaoka M. 2012. Bifidobacterial enzymes involved in the metabolism of human milk oligosaccharides. Adv Nutr 3:422s–429s. doi: 10.3945/an.111.001420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Good M, Sodhi CP, Yamaguchi Y, Jia H, Lu P, Fulton WB, Martin LY, Prindle T, Nino DF, Zhou Q, Ma C, Ozolek JA, Buck RH, Goehring KC, Hackam DJ. 2016. The human milk oligosaccharide 2′-fucosyllactose attenuates the severity of experimental necrotising enterocolitis by enhancing mesenteric perfusion in the neonatal intestine. Br J Nutr 116:1175–1187. doi: 10.1017/S0007114516002944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bode L. 2012. Human milk oligosaccharides: every baby needs a sugar mama. Glycobiology 22:1147–1162. doi: 10.1093/glycob/cws074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wada J, Ando T, Kiyohara M, Ashida H, Kitaoka M, Yamaguchi M, Kumagai H, Katayama T, Yamamoto K. 2008. Bifidobacterium bifidum lacto-N-biosidase, a critical enzyme for the degradation of human milk oligosaccharides with a type 1 structure. Appl Environ Microbiol 74:3996–4004. doi: 10.1128/AEM.00149-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moller PL, Jorgensen F, Hansen OC, Madsen SM, Stougaard P. 2001. Intra- and extracellular beta-galactosidases from Bifidobacterium bifidum and B. infantis: molecular cloning, heterologous expression, and comparative characterization. Appl Environ Microbiol 67:2276–2283. doi: 10.1128/AEM.67.5.2276-2283.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miwa M, Horimoto T, Kiyohara M, Katayama T, Kitaoka M, Ashida H, Yamamoto K. 2010. Cooperation of beta-galactosidase and beta-N-acetylhexosaminidase from bifidobacteria in assimilation of human milk oligosaccharides with type 2 structure. Glycobiology 20:1402–1409. doi: 10.1093/glycob/cwq101. [DOI] [PubMed] [Google Scholar]
- 16.Ashida H, Miyake A, Kiyohara M, Wada J, Yoshida E, Kumagai H, Katayama T, Yamamoto K. 2009. Two distinct alpha-l-fucosidases from Bifidobacterium bifidum are essential for the utilization of fucosylated milk oligosaccharides and glycoconjugates. Glycobiology 19:1010–1017. doi: 10.1093/glycob/cwp082. [DOI] [PubMed] [Google Scholar]
- 17.Kiyohara M, Tanigawa K, Chaiwangsri T, Katayama T, Ashida H, Yamamoto K. 2011. An exo-alpha-sialidase from bifidobacteria involved in the degradation of sialyloligosaccharides in human milk and intestinal glycoconjugates. Glycobiology 21:437–447. doi: 10.1093/glycob/cwq175. [DOI] [PubMed] [Google Scholar]
- 18.Wada J, Suzuki R, Fushinobu S, Kitaoka M, Wakagi T, Shoun H, Ashida H, Kumagai H, Katayama T, Yamamoto K. 2007. Purification, crystallization and preliminary X-ray analysis of the galacto-N-biose-/lacto-N-biose I-binding protein (GL-BP) of the ABC transporter from Bifidobacterium longum JCM1217. Acta Crystallogr Sect F Struct Biol Cryst Commun 63:751–753. doi: 10.1107/S1744309107036263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sakurama H, Kiyohara M, Wada J, Honda Y, Yamaguchi M, Fukiya S, Yokota A, Ashida H, Kumagai H, Kitaoka M, Yamamoto K, Katayama T. 2013. Lacto-N-biosidase encoded by a novel gene of Bifidobacterium longum subspecies longum shows unique substrate specificity and requires a designated chaperone for its active expression. J Biol Chem 288:25194–25206. doi: 10.1074/jbc.M113.484733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sela DA, Chapman J, Adeuya A, Kim JH, Chen F, Whitehead TR, Lapidus A, Rokhsar DS, Lebrilla CB, German JB, Price NP, Richardson PM, Mills DA. 2008. The genome sequence of Bifidobacterium longum subsp. infantis reveals adaptations for milk utilization within the infant microbiome. Proc Natl Acad Sci U S A 105:18964–18969. doi: 10.1073/pnas.0809584105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yoshida E, Sakurama H, Kiyohara M, Nakajima M, Kitaoka M, Ashida H, Hirose J, Katayama T, Yamamoto K, Kumagai H. 2012. Bifidobacterium longum subsp. infantis uses two different beta-galactosidases for selectively degrading type-1 and type-2 human milk oligosaccharides. Glycobiology 22:361–368. doi: 10.1093/glycob/cwr116. [DOI] [PubMed] [Google Scholar]
- 22.Garrido D, Ruiz-Moyano S, Mills DA. 2012. Release and utilization of N-acetyl-d-glucosamine from human milk oligosaccharides by Bifidobacterium longum subsp. infantis. Anaerobe 18:430–435. doi: 10.1016/j.anaerobe.2012.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xiao JZ, Takahashi S, Nishimoto M, Odamaki T, Yaeshima T, Iwatsuki K, Kitaoka M. 2010. Distribution of in vitro fermentation ability of lacto-N-biose I, a major building block of human milk oligosaccharides, in bifidobacterial strains. Appl Environ Microbiol 76:54–59. doi: 10.1128/AEM.01683-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sela DA, Li Y, Lerno L, Wu S, Marcobal AM, German JB, Chen X, Lebrilla CB, Mills DA. 2011. An infant-associated bacterial commensal utilizes breast milk sialyloligosaccharides. J Biol Chem 286:11909–11918. doi: 10.1074/jbc.M110.193359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sela DA, Garrido D, Lerno L, Wu S, Tan K, Eom H-J, Joachimiak A, Lebrilla CB, Mills DA. 2012. Bifidobacterium longum subsp. infantis ATCC 15697 α-fucosidases are active on fucosylated human milk oligosaccharides. Appl Environ Microbiol 78:795–803. doi: 10.1128/AEM.06762-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bunesova V, Lacroix C, Schwab C. 2016. Fucosyllactose and l-fucose utilization of infant Bifidobacterium longum and Bifidobacterium kashiwanohense. BMC Microbiol 16:248. doi: 10.1186/s12866-016-0867-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.James K, Motherway MO, Bottacini F, van Sinderen D. 2016. Bifidobacterium breve UCC2003 metabolises the human milk oligosaccharides lacto-N-tetraose and lacto-N-neo-tetraose through overlapping, yet distinct pathways. Sci Rep 6:38560. doi: 10.1038/srep38560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Egan M, Motherway MO, Kilcoyne M, Kane M, Joshi L, Ventura M, van Sinderen D. 2014. Cross-feeding by Bifidobacterium breve UCC2003 during co-cultivation with Bifidobacterium bifidum PRL2010 in a mucin-based medium. BMC Microbiol 14:282. doi: 10.1186/s12866-014-0282-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Egan M, O'Connell Motherway M, Ventura M, van Sinderen D. 2014. Metabolism of sialic acid by Bifidobacterium breve UCC2003. Appl Environ Microbiol 80:4414–4426. doi: 10.1128/AEM.01114-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Egan M, Jiang H, O'Connell Motherway M, Oscarson S, van Sinderen D. 2016. Glycosulfatase-encoding gene cluster in Bifidobacterium breve UCC2003. Appl Environ Microbiol 82:6611–6623. doi: 10.1128/AEM.02022-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Titgemeyer F, Reizer J, Reizer A, Saier MH Jr. 1994. Evolutionary relationships between sugar kinases and transcriptional repressors in bacteria. Microbiology 140(Part 9):2349–2354. doi: 10.1099/13500872-140-9-2349. [DOI] [PubMed] [Google Scholar]
- 32.O'Connell Motherway M, Fitzgerald GF, Neirynck S, Ryan S, Steidler L, van Sinderen D. 2008. Characterization of ApuB, an extracellular type II amylopullulanase from Bifidobacterium breve UCC2003. Appl Environ Microbiol 74:6271–6279. doi: 10.1128/AEM.01169-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ruiz L, Motherway MO, Lanigan N, van Sinderen D. 2013. Transposon mutagenesis in Bifidobacterium breve: construction and characterization of a Tn5 transposon mutant library for Bifidobacterium breve UCC2003. PLoS One 8:e64699. doi: 10.1371/journal.pone.0064699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ruiz L, Zomer A, O'Connell-Motherway M, van Sinderen D, Margolles A. 2012. Discovering novel bile protection systems in Bifidobacterium breve UCC2003 through functional genomics. Appl Environ Microbiol 78:1123–1131. doi: 10.1128/AEM.06060-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hawley DK, McClure WR. 1983. Compilation and analysis of Escherichia coli promoter DNA sequences. Nucleic Acids Res 11:2237–2255. doi: 10.1093/nar/11.8.2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bottacini F, Zomer A, Milani C, Ferrario C, Lugli GA, Egan M, Ventura M, van Sinderen D. 2017. Global transcriptional landscape and promoter mapping of the gut commensal Bifidobacterium breve UCC2003. BMC Genomics 18:991. doi: 10.1186/s12864-017-4387-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pokusaeva K, O'Connell-Motherway M, Zomer A, Macsharry J, Fitzgerald GF, van Sinderen D. 2011. Cellodextrin utilization by Bifidobacterium breve UCC2003. Appl Environ Microbiol 77:1681–1690. doi: 10.1128/AEM.01786-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.O'Connell KJ, Motherway MO, Liedtke A, Fitzgerald GF, Ross RP, Stanton C, Zomer A, van Sinderen D. 2014. Transcription of two adjacent carbohydrate utilization gene clusters in Bifidobacterium breve UCC2003 is controlled by LacI- and repressor open reading frame kinase (ROK)-type regulators. Appl Environ Microbiol 80:3604–3614. doi: 10.1128/AEM.00130-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Egan M, O'Connell Motherway M, van Sinderen D. 2015. A GntR-type transcriptional repressor controls sialic acid utilization in Bifidobacterium breve UCC2003. FEMS Microbiol Lett 362:1–9. doi: 10.1093/femsle/fnu056. [DOI] [PubMed] [Google Scholar]
- 40.Ravcheev DA, Khoroshkin MS, Laikova ON, Tsoy OV, Sernova NV, Petrova SA, Rakhmaninova AB, Novichkov PS, Gelfand MS, Rodionov DA. 2014. Comparative genomics and evolution of regulons of the LacI-family transcription factors. Front Microbiol 5:294. doi: 10.3389/fmicb.2014.00294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gilbert W, Maxam A. 1973. The nucleotide sequence of the lac operator. Proc Natl Acad Sci U S A 70:3581–3584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.O'Connell Motherway M, Fitzgerald GF, van Sinderen D. 2011. Metabolism of a plant derived galactose-containing polysaccharide by Bifidobacterium breve UCC2003. Microb Biotechnol 4:403–416. doi: 10.1111/j.1751-7915.2010.00218.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pokusaeva K, Neves AR, Zomer A, O'Connell-Motherway M, MacSharry J, Curley P, Fitzgerald GF, van Sinderen D. 2010. Ribose utilization by the human commensal Bifidobacterium breve UCC2003. Microb Biotechnol 3:311–323. doi: 10.1111/j.1751-7915.2009.00152.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Matthews KS, Nichols JC. 1998. Lactose repressor protein: functional properties and structure. Prog Nucleic Acid Res Mol Biol 58:127–164. doi: 10.1016/S0079-6603(08)60035-5. [DOI] [PubMed] [Google Scholar]
- 45.Lewis M. 2005. The lac repressor. C R Biol 328:521–548. doi: 10.1016/j.crvi.2005.04.004. [DOI] [PubMed] [Google Scholar]
- 46.Plumbridge J. 2001. DNA binding sites for the Mlc and NagC proteins: regulation of nagE, encoding the N-acetylglucosamine-specific transporter in Escherichia coli. Nucleic Acids Res 29:506–514. doi: 10.1093/nar/29.2.506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Brechemier-Baey D, Dominguez-Ramirez L, Oberto J, Plumbridge J. 2015. Operator recognition by the ROK transcription factor family members, NagC and Mlc. Nucleic Acids Res 43:361–372. doi: 10.1093/nar/gku1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Khoroshkin MS, Leyn SA, Van Sinderen D, Rodionov DA. 2016. Transcriptional regulation of carbohydrate utilization pathways in the Bifidobacterium genus. Front Microbiol 7:120. doi: 10.3389/fmicb.2016.00120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Weickert MJ, Adhya S. 1992. A family of bacterial regulators homologous to Gal and Lac repressors. J Biol Chem 267:15869–15874. [PubMed] [Google Scholar]
- 50.Weickert MJ, Adhya S. 1993. The galactose regulon of Escherichia coli. Mol Microbiol 10:245–251. doi: 10.1111/j.1365-2958.1993.tb01950.x. [DOI] [PubMed] [Google Scholar]
- 51.O'Connell Motherway M, Kinsella M, Fitzgerald GF, van Sinderen D. 2013. Transcriptional and functional characterization of genetic elements involved in galacto-oligosaccharide utilization by Bifidobacterium breve UCC2003. Microb Biotechnol 6:67–79. doi: 10.1111/1751-7915.12011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Barboza M, Sela DA, Pirim C, LoCascio RG, Freeman SL, German JB, Mills DA, Lebrilla CB. 2009. Glycoprofiling bifidobacterial consumption of galacto-oligosaccharides by mass spectrometry reveals strain-specific, preferential consumption of glycans. Appl Environ Microbiol 75:7319–7325. doi: 10.1128/AEM.00842-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Coulier L, Timmermans J, Bas R, Van Den Dool R, Haaksman I, Klarenbeek B, Slaghek T, Van Dongen W. 2009. In-depth characterization of prebiotic galacto-oligosaccharides by a combination of analytical techniques. J Agric Food Chem 57:8488–8495. doi: 10.1021/jf902549e. [DOI] [PubMed] [Google Scholar]
- 54.Pokusaeva K, Fitzgerald GF, van Sinderen D. 2011. Carbohydrate metabolism in bifidobacteria. Genes Nutr 6:285–306. doi: 10.1007/s12263-010-0206-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.de Man JC, Rogosa M, Sharpe ME. 1960. A medium for the cultivation of 688 lactobacilli. J Appl Bacteriol 23:130–135. doi: 10.1111/j.1365-2672.1960.tb00188.x. [DOI] [Google Scholar]
- 56.Watson D, O'Connell Motherway M, Schoterman MH, van Neerven RJ, Nauta A, van Sinderen D. 2013. Selective carbohydrate utilization by lactobacilli and bifidobacteria. J Appl Microbiol 114:1132–1146. doi: 10.1111/jam.12105. [DOI] [PubMed] [Google Scholar]
- 57.Terzaghi BE, Sandine WE. 1975. Improved medium for lactic streptococci and their bacteriophages. Appl Microbiol 29:807–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 59.Rutherford K, Parkhill J, Crook J, Horsnell T, Rice P, Rajandream MA, Barrell B. 2000. Artemis: sequence visualization and annotation. Bioinformatics 16:944–945. doi: 10.1093/bioinformatics/16.10.944. [DOI] [PubMed] [Google Scholar]
- 60.O'Connell Motherway M, Zomer A, Leahy SC, Reunanen J, Bottacini F, Claesson MJ, O'Brien F, Flynn K, Casey PG, Moreno Munoz JA, Kearney B, Houston AM, O'Mahony C, Higgins DG, Shanahan F, Palva A, de Vos WM, Fitzgerald GF, Ventura M, O'Toole PW, van Sinderen D. 2011. Functional genome analysis of Bifidobacterium breve UCC2003 reveals type IVb tight adherence (Tad) pili as an essential and conserved host-colonization factor. Proc Natl Acad Sci U S A 108:11217–11222. doi: 10.1073/pnas.1105380108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol 215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 62.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.O'Riordan K, Fitzgerald GF. 1999. Molecular characterisation of a 5.75-kb cryptic plasmid from Bifidobacterium breve NCFB 2258 and determination of mode of replication. FEMS Microbiol Lett 174:285–294. doi: 10.1111/j.1574-6968.1999.tb13581.x. [DOI] [PubMed] [Google Scholar]
- 64.O'Connell Motherway M, O'Driscoll J, Fitzgerald GF, Van Sinderen D. 2009. Overcoming the restriction barrier to plasmid transformation and targeted mutagenesis in Bifidobacterium breve UCC2003. Microb Biotechnol 2:321–332. doi: 10.1111/j.1751-7915.2008.00071.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Maze A, O'Connell-Motherway M, Fitzgerald GF, Deutscher J, van Sinderen D. 2007. Identification and characterization of a fructose phosphotransferase system in Bifidobacterium breve UCC2003. Appl Environ Microbiol 73:545–553. doi: 10.1128/AEM.01496-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Law J, Buist G, Haandrikman A, Kok J, Venema G, Leenhouts K. 1995. A system to generate chromosomal mutations in Lactococcus lactis which allows fast analysis of targeted genes. J Bacteriol 177:7011–7018. doi: 10.1128/jb.177.24.7011-7018.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Egan M, O'Connell Motherway M, Ventura M, van Sinderen D. 9 May 2014. Metabolism of sialic acid by Bifidobacterium breve UCC2003. Appl Environ Microbiol doi: 10.1128/aem.01114-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zomer A, Fernandez M, Kearney B, Fitzgerald GF, Ventura M, van Sinderen D. 2009. An interactive regulatory network controls stress response in Bifidobacterium breve UCC2003. J Bacteriol 191:7039–7049. doi: 10.1128/JB.00897-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Garcia de la Nava J, Santaella DF, Cuenca Alba J, Maria Carazo J, Trelles O, Pascual-Montano A. 2003. Engene: the processing and exploratory analysis of gene expression data. Bioinformatics 19:657–658. doi: 10.1093/bioinformatics/btg028. [DOI] [PubMed] [Google Scholar]
- 70.van Hijum S, de Jong A, Baerends R, Karsens H, Kramer N, Larsen R, den Hengst C, Albers C, Kok J, Kuipers O. 2005. A generally applicable validation scheme for the assessment of factors involved in reproducibility and quality of DNA-microarray data. BMC Genomics 6:77. doi: 10.1186/1471-2164-6-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.van Hijum SA, Garcia de la Nava J, Trelles O, Kok J, Kuipers OP. 2003. MicroPreP: a cDNA microarray data pre-processing framework. Appl Bioinformatics 2:241–244. [PubMed] [Google Scholar]
- 72.Long AD, Mangalam HJ, Chan BY, Tolleri L, Hatfield GW, Baldi P. 2001. Improved statistical inference from DNA microarray data using analysis of variance and a Bayesian statistical framework. Analysis of global gene expression in Escherichia coli K12. J Biol Chem 276:19937–19944. doi: 10.1074/jbc.M010192200. [DOI] [PubMed] [Google Scholar]
- 73.Mierau I, Kleerebezem M. 2005. 10 years of the nisin-controlled gene expression system (NICE) in Lactococcus lactis. Appl Microbiol Biotechnol 68:705–717. doi: 10.1007/s00253-005-0107-6. [DOI] [PubMed] [Google Scholar]
- 74.O'Connell KJ, O'Connell Motherway M, O'Callaghan J, Fitzgerald GF, Ross RP, Ventura M, Stanton C, van Sinderen D. 2013. Metabolism of four alpha-glycosidic linkage-containing oligosaccharides by Bifidobacterium breve UCC2003. Appl Environ Microbiol 79:6280–6292. doi: 10.1128/AEM.01775-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.de Ruyter PG, Kuipers OP, de Vos WM. 1996. Controlled gene expression systems for Lactococcus lactis with the food-grade inducer nisin. Appl Environ Microbiol 62:3662–3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hamoen LW, Van Werkhoven AF, Bijlsma JJ, Dubnau D, Venema G. 1998. The competence transcription factor of Bacillus subtilis recognizes short A/T-rich sequences arranged in a unique, flexible pattern along the DNA helix. Genes Dev 12:1539–1550. doi: 10.1101/gad.12.10.1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kuipers OP, Beerthuyzen MM, Siezen RJ, De Vos WM. 1993. Characterization of the nisin gene cluster nisABTCIPR of Lactococcus lactis. Requirement of expression of the nisA and nisI genes for development of immunity. Eur J Biochem 216:281–291. doi: 10.1111/j.1432-1033.1993.tb18143.x. [DOI] [PubMed] [Google Scholar]
- 78.Ventura M, Zink R, Fitzgerald GF, van Sinderen D. 2005. Gene structure and transcriptional organization of the dnaK operon of Bifidobacterium breve UCC 2003 and application of the operon in bifidobacterial tracing. Appl Environ Microbiol 71:487–500. doi: 10.1128/AEM.71.1.487-500.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bailey TL, Elkan C. 1994. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc Int Conf Intell Syst Mol Biol 2:28–36. [PubMed] [Google Scholar]
- 80.Crooks GE, Hon G, Chandonia JM, Brenner SE. 2004. WebLogo: a sequence logo generator. Genome Res 14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Schneider TD, Stephens RM. 1990. Sequence logos: a new way to display consensus sequences. Nucleic Acids Res 18:6097–6100. doi: 10.1093/nar/18.20.6097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Álvarez-Martín P, O'Connell-Motherway M, van Sinderen D, Mayo B. 2007. Functional analysis of the pBC1 replicon from Bifidobacterium catenulatum L48. Appl Microbiol Biotechnol 76:1395–1402. doi: 10.1007/s00253-007-1115-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.