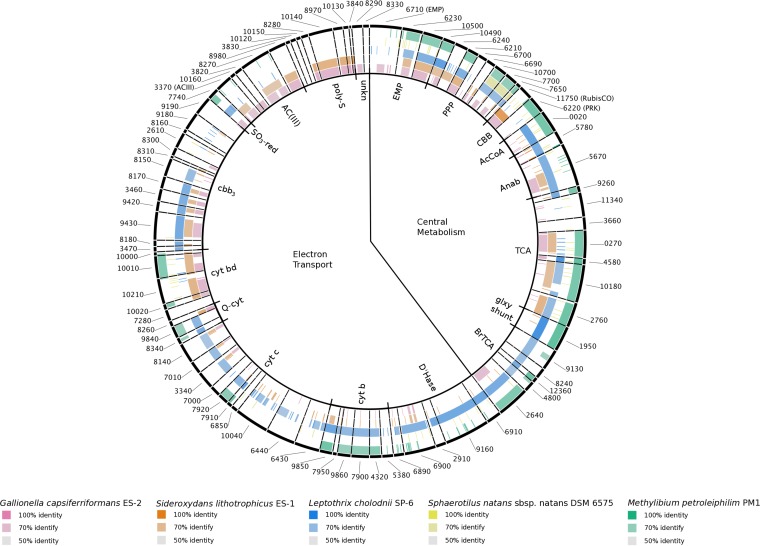
FIG 3.
Genes coding for proteins putatively involved in central metabolism and the electron transport chain. Protein sequences from L. ochracea's partial genome are compared to those in other taxa generated using the BLAST Ring Generator. L. ochracea is compared to Gallionella capsiferriformans ES-2 (rose), Sideroxydans lithotrophicus ES-1 (orange), L. cholodnii SP-6 (blue), Sphaerotilus natans (yellow), and Methylibium petroleiphilum PM1 (green). The intensity of color describes the percent sequence identity, and these are grouped clockwise around the ring starting at the top with central metabolism, with the Emden-Meyerhof-Parnas (EMP) pathway, pentose phosphate pathway (PPP), Calvin-Benson-Bassham (CBB) cycle, acetyl coenzyme A (AcCoA) connector, anapleurotic anabolic (Anab) reactions, tricarboxylic acid (TCA) cycle, horseshoe or branched TCA (BrTCA) cycle, and glyoxylate shunt (glxy_shnt) for central metabolism. The putative electron transport chain enzymes include dehydrogenases (D'Hase), cytochrome b subunits (b/b6), cytochrome c (cyt c), ubiquinol-cytochrome c reductase (Q-cytc), cytochrome bd (cyt bd), cytochrome cbb3 (cbb3) sulfite reductase (SO3-red), alternative complex III (ACIII), polysulfide reductase (poly-S), and proteins of unknown function.