FIG 5.
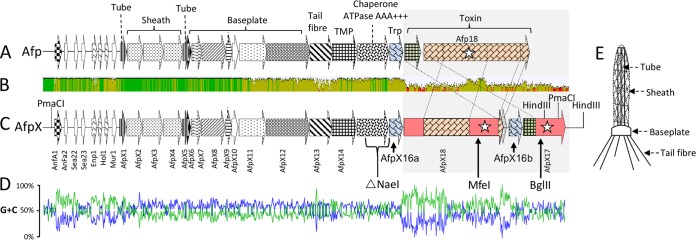
(A and C) Schematic representations of the S. entomophila Afp (A) and S. proteamaculans AGR96X AfpX (C) gene clusters. ORFs are denoted by arrows, with their designations listed vertically below. Amino acid regions with high similarity are indicated by similar shading (see Table 3 for the percent amino acid similarity/identity of the respective AfpX proteins). Stars indicate ORFs for which the translated product shows similarity to a documented virulence factor. Predicted and ascribed protein domains are listed above the S. entomophila afp gene cluster in panel A. (TMP denotes tape measure protein and Trp denotes tail-length termination protein.) The NaeI region deleted in the AGR96XΔAFP1516a deletion variant (Table 4) is indicated below the ORFs corresponding to AfpX15 and AfpX16 in panel C. The MfeI and BglII sites used to insert markers to generate AGR96X::18 and AGR96X::17 (Table 4) are indicted in panel C. (B) Mean pairwise afp and afpX nucleotide identities over all pairs in the column. Green, 100% identity; olive, ≥30 to 99% identity; red, <30% identity. Angled dashed lines denote shared areas of amino acid similarity between Afp17 and Afp18 and their AfpX orthologs. (D) G+C skew plot of nucleotide identity shared between Afp- and AfpX-coding regions. (E) Schematic representation of Afp with the structural parts, namely, sheath, tube, tail fibers, and baseplate, designated.