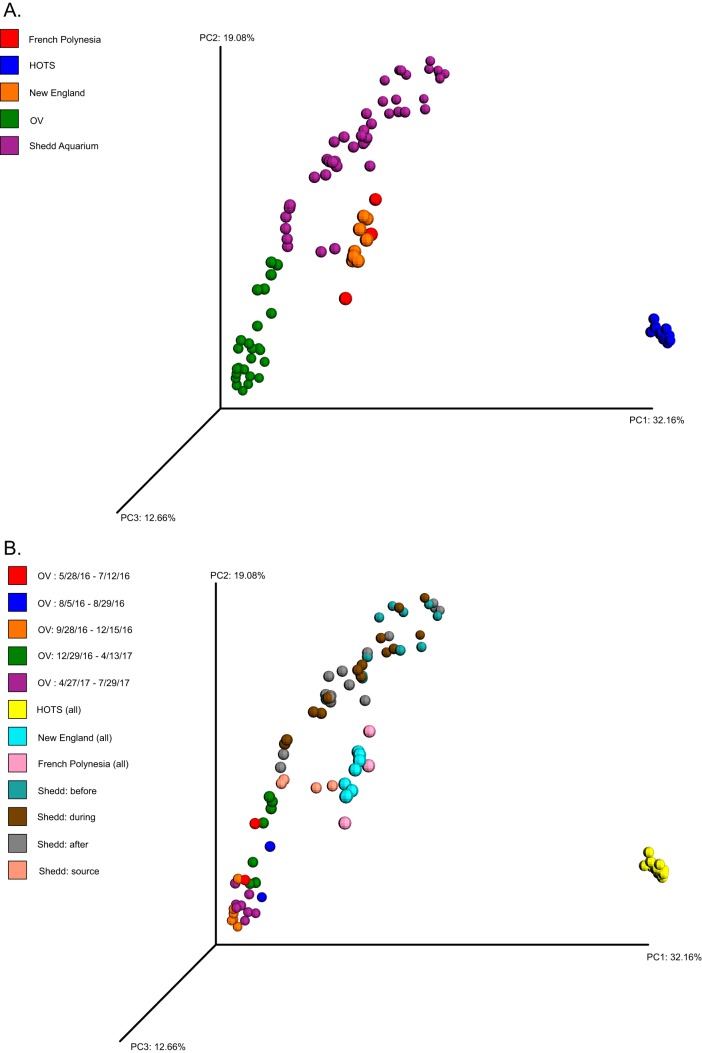
FIG 4.
Principal-component analysis comparing microbiome compositions among samples from the Ocean Voyager and the Shedd Aquarium (aquarium water samples) as well as Station ALOHA (HOTS) (25 m), French Polynesia (<1 m), and New England (<1 m) (natural seawater samples). Clustering is based on the weighted UniFrac metric generated from the 16S rRNA gene amplicon data of all samples. (A) Samples are colored by environment. (B) The aquarium samples are further classified by collection time period (OV) or, for the Shedd Aquarium data, by experiment stage (before, during, and after a 90% water change; the data set also contains a sample of the replacement water used for the water change). HOTS, French Polynesia, and New England samples are colored based only on environment, as in panel A.