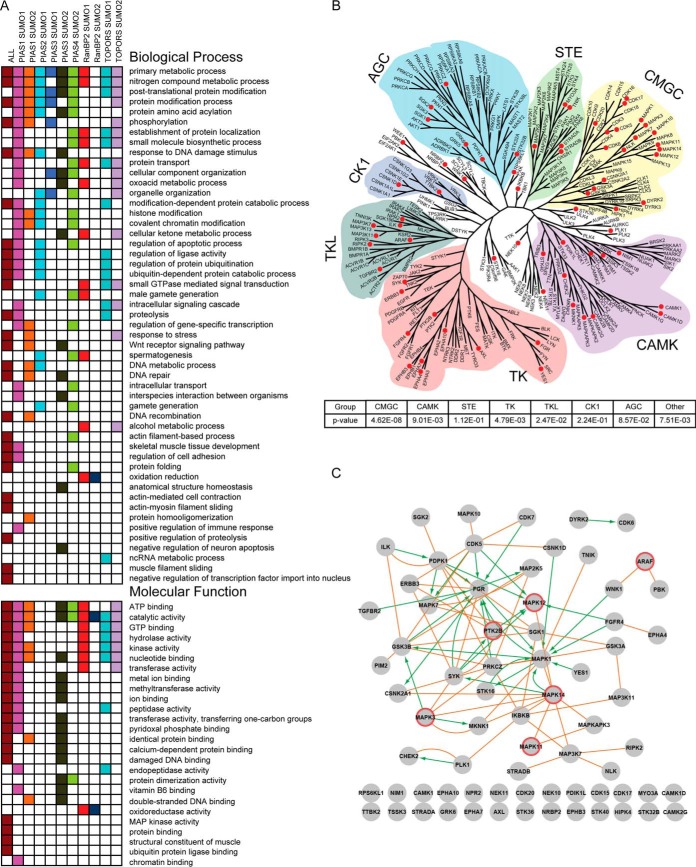
Fig. 3.
Biological Classification of SUMOylated proteins. A, Gene Ontology analysis of significant biological processes and molecular functions. Overrepresented (p < 0.05) biological process and molecular function GO categories were identified. The composite data set and each E3 ligase/SUMO pairing were analyzed separately. B, Phylogenetic kinase tree overlaid with SUMOylation enrichment the amino acid sequences of kinase domain of all human kinase proteins were collected to build the phylogenetic tree with Mega 5, a program designed to construct phylogenetic trees from aligned sequences. Kinase families are designated by distinct color and SUMOylated kinases are indicated by red circles. C, Kinase SUMOylation substrates in PPI and KSR network. Protein-protein interaction analysis was performed on the subset of kinases identified as SUMO substrates in our array-based assays (orange edges). Kinase substrate relationships (KSRs) are represented by green directional arrows. Kinases encircled in red were selected for validation studies.