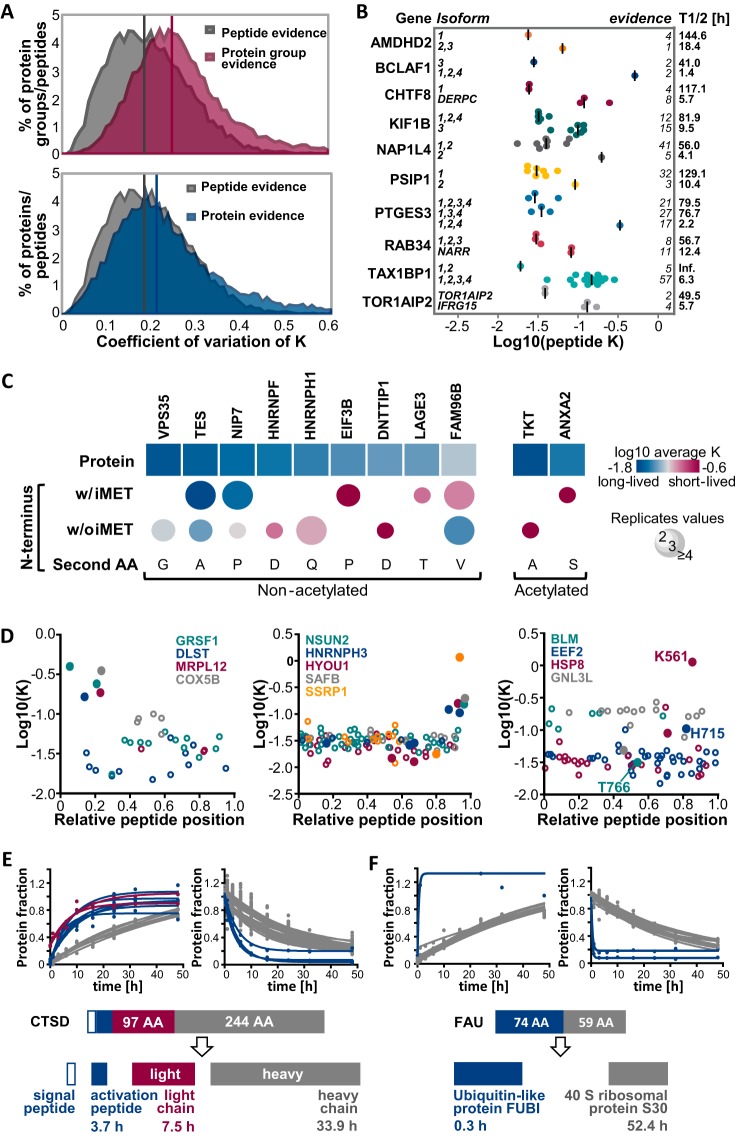
Fig. 6.
Analysis of proteoform resolved protein turnover. A, Distributions of coefficients of variation of turnover rates across spectra are displayed for peptides and proteins. The upper panel shows the distribution of CVs of all spectra for the same protein group (irrespective of the number of proteins or protein isoforms in each group). The bottom panel shows the CV distribution of all spectra for protein groups that only contain a single protein. Medians are indicated by vertical lines in the corresponding color. B, Scatter dot plots depict the distributions of turnover rate constants for peptides of different protein isoforms. Peptides corresponding to a gene share the same color. Median peptide rates across replicates are shown as vertical lines. C, Turnover rates are shown for different modified N-terminal peptides and corresponding proteins. Only N termini and proteins with statistically significantly different rates are displayed (see also supplemental Fig. S6C). D, Scatter plots illustrate peptide turnover rates as a function of the relative position within the protein sequence. Zero denotes the N terminus and 1 denotes the C terminus of a protein. Peptides from each protein are denoted by the same color whereas closed circles denote peptides that exhibit significantly different turnover compared with the rate of the whole protein (see also supplemental Fig. S6B). The left panel shows examples for mitochondrial proteins in which the N-terminal transit peptides shows a higher turnover than other peptides of the same protein. The middle panel shows a similar analysis but for proteins with higher turnover C-terminal peptides. The right panel shows examples for proteins in which one peptide displayed a strong difference in turnover compared with other peptides of the same protein which often but not always encompass known modification sites. E, Fractional peptide labeling is depicted for cathepsin D (CTSD), a protein that is proteolytically processed into a signal peptide, an activation peptide (blue circles), a light chain (red circles) and a heavy chain (gray circles). F, Fractional peptide labeling is depicted for the fusion protein FAU. Peptides representing the Ubiquitin-like protein FUBI are shown in blue, peptides from the 40S ribosomal protein S30 are shown in gray.