Abstract
Background
The tick-borne protozoan parasite Theileria parva causes a usually fatal cattle disease known as East Coast fever in sub-Saharan Africa, with devastating consequences for poor small-holder farmers. Immunity to T. parva, believed to be mediated by a cytotoxic T lymphocyte (CTL) response, is induced following natural infection and after vaccination with a live vaccine, known as the Infection and Treatment Method (ITM). The most commonly used version of ITM is a combination of parasites derived from three isolates (Muguga, Kiambu 5 and Serengeti-transformed), known as the “Muguga cocktail”. The use of a vaccine comprising several strains is believed to be required to induce a broad immune response effective against field challenge. In this study we investigated whether immunization with the Muguga cocktail induces a broader CTL response than immunization with a single strain (Muguga).
Results
Four MHC haplotype-matched pairs of cattle were immunized with either the trivalent Muguga cocktail or the single Muguga strain. CTL specificity was assessed on a panel of five different strains, and clonal responses to these strains were also assessed in one of the MHC-matched pairs. We did not find evidence for a broader CTL response in animals immunized with the Muguga cocktail compared to those immunized with the Muguga strain alone, in either the bulk or clonal CTL analyses. This was supported by an in vivo trial in which all vaccinated animals survived challenge with a lethal dose of the Muguga cocktail vaccine stabilate.
Conclusion
We did not observe any substantial differences in the immunity generated from animals immunized with either Muguga alone or the Muguga cocktail in the animals tested here, corroborating earlier results showing limited antigenic diversity in the Muguga cocktail. These results may warrant further field studies using single T. parva strains as future vaccine candidates.
Keywords: Theileria parva, Live vaccine, Cytotoxic T cells, Immunity, Strain specificity
Background
Theileria parva is a tick-borne protozoan parasite which causes an acute and usually fatal cattle disease, known as East Coast fever, in eastern, central and southern Africa. The parasite infects bovine lymphocytes, which subsequently undergo blast transformation and rapid multiplication [1]. In susceptible animals, this usually results in overwhelming parasitosis and death within 2 to 4 weeks of infection.
Cattle which recover from natural infection can develop a strong immunity to subsequent challenge. This has been exploited to develop a vaccination procedure known as the “Infection and Treatment Method” (ITM) in which live sporozoites are administered simultaneously with oxytetracycline. The main protective mechanism in both vaccinated and naturally recovered animals is believed to be CD8+ cytotoxic T lymphocyte (CTL) killing of infected lymphocytes. Thus, adoptive transfer of CD8+ cells from immunized animals has been demonstrated to protect naïve animals from challenge [2]. Furthermore, it has been shown that the time point of recovery correlates with a peak of CD8+ cells in the blood of the infected animals [3].
Early experiments in the development of the ITM vaccine revealed the presence of strain specificity through infection and challenge trials where animals were immunized with one strain and challenged with a heterologous strain. While good protection was often obtained following the inoculation of single strains, it did not always extend to heterologous challenge [4]. It was subsequently shown that a combination of three strains (Muguga, Serengeti-transformed and Kiambu 5) provided better protection than single strains. The mixture, known as the “Muguga cocktail”, is the basis of a commercial ITM vaccine which appears to provide broad protection against T. parva in the field, so far best explored in Tanzania [5, 6]. It should be noted that in experiments involving single isolates, some cross-protection was observed with a notable exception being if animals vaccinated with the Muguga strain or Kilifi strain were challenged with Marikebuni [4, 7, 8], suggesting that Marikebuni is antigenically quite distinct.
Strain specificity has also been observed in in vitro CTL assays and reflects the in vivo immune status [3, 9–11]. A possible explanation for the strain specificity of the CTL response to T. parva is based on two phenomena – immunodominance and antigenic diversity. Immunodominance, where the immune response is directed to a very limited number of antigens in individual animals, is commonly observed in viral infections [12–15] and also in other infectious diseases and cancers [16–18]. In T. parva infections, indications of immunodominance came with initial analyses of the immune response where, in some cases, it was possible to show that CTL restriction was mediated by a single class I MHC molecule [19, 20] despite the presence of many potential epitopes expressed by the parasite genome, which is predicted to encode 4034 genes [21, 22], and a T cell receptor (TCR) repertoire capable of reacting to a wide variety of epitopes. More recently, with the discovery of CTL antigens [23], a study was performed to shed light on the issue of immunodominance [24]. Findings from this work suggest that one or a few MHC alleles in individual animals govern the specificity of the immune response elicited by infection or vaccination and are crucial for the outcome of a later challenge with genotypically different parasites.
The most plausible explanation for the broad protection offered by the Muguga cocktail, is that the mixture of the parasites in the cocktail, provides a more diverse set of antigens which potentially can induce a CTL response of broader antigenic specificity than inoculation with any of the individual components or single strains, and thus provide better protection against heterologous parasites encountered in the field. To test this hypothesis, we compared the specificities for different parasite strains of CTL induced in animals immunized by ITM with the Muguga cocktail with those generated by the Muguga stabilate alone. To minimize any effect of the MHC background in individual animals, which is known to influence the selection of antigens recognized by the CTLs, the responses were compared in MHC haploidentical pairs of cattle. Analysis of the CTL response at the clonal level was also undertaken for one haplotype matched pair. All haploidentical pairs of cattle were challenged with the Muguga cocktail to uncover any antigenic differences in the in vivo CTL response.
Methods
Animals and MHC class I typing
All animal experiments were reviewed and approved by the Institutional Animal Care and Use Committee at International Livestock Research Institute (ILRI). Eight Bos taurus cattle (five Friesian and three Ayrshire) were bought from farms in the Nyeri area in Kenya. They were screened free for tickborne diseases including T. parva, and BoLA typed using a combination of serology (ELISA using antibodies defining particular MHC haplotypes), IFNγ ELISPOT assay using PBMC from the cattle pulsed with Tp1214–224 and a peptide-specific CTL line, and by PCR using haplotype specific primers followed by sequencing, essentially as described before [25]. Four haploidentical pairs of the following MHC haplotypes were selected for the study: A10/A12, A12/A14, A15/ A18, A11/A15 (Table 1). Animals were kept in standard pens and were fed normally. At the end of the experiment, animals were returned to the farm and eventually slaughtered for meat. Two control animals, which developed disease, were euthanized for humane reasons using an overdose of Euthatal (Pentobarbital sodium, 200 mg/ml), 1 ml Euthatal per 1.4 kg body weight, given intravenously, after restraining the animals.
Table 1.
Cattle used in the study
| Calf | Breed | MHC Class 1 Haplotype |
Alleles | Immunization |
|---|---|---|---|---|
| BG033 | Friesian | A10/A12 | N*00201, N*01901 | Muguga (3308) |
| BG042 | Friesian | A10/A12 | N*00201, N*01901 | Muguga cocktail (0801) |
| BG053 | Ayrshire | A12/A14 | N*01901, N*02301 | Muguga (3308) |
| BG051 | Ayrshire | A12/A14 | N*01901, N*02301 | Muguga cocktail (0801) |
| BG052 | Friesian | A15/A18 | N*00901, N*01302 | Muguga (3308) |
| BG056 | Friesian | A15/A18 | N*00901, N*01302 | Muguga cocktail (0801) |
| BH055 | Ayrshire | A11/A15 | N*01802, N*00902 | Muguga (3308) |
| BH047 | Friesian | A11/A15 | N*01802, N*00902 | Muguga cocktail (0801) |
The breed, MHC Class I haplotype and associated MHC alleles of the cattle included in the study are shown. Within each of the haplotype matched pairs of cattle, one was immunized with the single T. parva strain Muguga 3308 and the other was immunized with the Muguga cocktail 0801 using ITM
Immunization
Each of the haplotype-matched pairs of cattle was inoculated subcutaneously in front and below the right ear with either the Muguga stabilate 3308 or the Muguga cocktail vaccine stabilate ILRI0801 [26] and treated immediately with long acting oxytetracyclin.
Parasitized cell lines
Cell lines infected with T. parva were established by infection of autologous PBMC in vitro with sporozoites as described previously [27]. The sporozoites were from the cloned stabilates Marikebuni 3292, Muguga 3308, Boleni 3230, Uganda 3645, derived from 3569 [28], and Mariakani 3212 (unpublished). In addition, cell lines were established using sporozoites of the ILRI0801 reference stabilates: Muguga (4230), Serengeti (4229), Kiambu 5 (4228). These are stabilates of the individual Muguga cocktail components made from the same production ticks as the ILRI0801 vaccine [26].
Generation of CTL
CTL bulk cultures were generated and maintained in RPMI 1640 (Sigma-Aldrich, St.Louis, MO, USA) supplemented with 10% FBS (Thermo Fisher Scientific, Waltham, MA, USA), 2 mM L-glutamin (Sigma-Aldrich, St.Louis, MO, USA), 50 μM 2-mercaptoethanol (Sigma-Aldrich, St.Louis, MO, USA), 100 IU of penicillin/ml (Sigma-Aldrich, St.Louis, MO, USA), 100 μg of streptomycin/ml (Sigma-Aldrich, St.Louis, MO, USA), 50 μg of gentamicin/ml, (Sigma-Aldrich, St.Louis, MO, USA) 10% TCGF (Conditioned media from ConA blasts).
CTL were generated essentially as described [27, 29]. Briefly, PBMC were re-stimulated three times with irradiated autologous T. parva-infected cell lines. CTL from animals immunized with the vaccine were generated by stimulating with equal fractions of three cell lines infected with one of the three vaccine reference stabilates. CTL from the Muguga- immunized animals were stimulated with the Muguga-infected cell line only. The remainder of the procedure was as described previously [27, 29].
Generation of CD8+ CTL clones
Clones were generated by purifying CD8+ cells after two restimulations as described above. Enrichment of CD8+ cells was achieved by incubating the CTL line for 30 min with mouse-anti-bovine CD8 mAb ILA105 (ILRI) diluted 1:500 in PBS + 2% FBS. Cells were washed twice in PBS + 2%FBS, labeled with magnetic beads attached to goat-anti-mIgG (Miltenyi Biotec, Bergish Gladbach, Germany) and purified according to the protocol provided by the manufacturer. Purified CD8+ cells were then seeded by limiting dilutions in 96-well plates using 2 × 104 irradiated PBMC as filler cells as previously described [27].
CTL lines and clones were generated and maintained in RPMI 1640 medium (Sigma-Aldrich, St. Louis, MO, USA) supplemented with 10% FBS (Thermo Fisher Scientific, Waltham, MA, USA), 2 mM L-glutamine, 50 μM 2-mercaptoethanol, 100 IU/ml of penicillin, 100 μg/ml of streptomycin, 50 μg/ml of gentamicin, (all from Sigma-Aldrich, St. Louis, MO, USA) and 10% inactivated ConA supernatant (conditioned media from ConA blasts).
Cytotoxicity assay
A standard 4 h release assay using 51Cr-labeled target cells was used to measure cytotoxicity. 51Cr was obtained from American Radiolabeled Chemicals, Inc., St. Louis, MO, USA. Supernatants were counted using Lumaplates (PerkinElmer, Waltham, MA, USA) in a TopCounter (PerkinElmer, Waltham, MA, USA). The cytotoxicity was calculated as: (experimental release-spontaneous release/total release-spontaneous release). Target cells were either T. parva-infected autologous PBMC using MHC-mismatched T. parva-infected cell lines as controls. In one case (BH055), autologous uninfected PBMC were used as control. Each CTL was tested in dilutions using a fixed number of target cells. Each dilution was tested in triplicate. Cytotoxicity at an effector:target ratio of 10:1 was used to compare between CTL derived from different animals and between different target cells.
Challenge experiment
Cattle were challenged with the Muguga cocktail vaccine stabilate ILRI0801, without oxytetracycline. The vaccine (2 ml neat) was injected subcutaneously in front of and below the right ear. Animals were monitored for the required clinical parameters to determine the severity of disease according to the Rowlands index [30].
Results
Similar strain specificities of CTL from haplotype-matched animals immunized by ITM with either T. parva Muguga or the Muguga cocktail
Eight cattle comprising four haploidentical pairs were immunized by ITM with either the Muguga strain (3308) or the trivalent Muguga cocktail (ILRI0801), as detailed in Table 1.
CTL were generated from all animals and tested against autologous cell lines infected with five cloned sporozoite stabilates and an MHC-mismatched infected cell line as control. Figure 1 shows an example of the results for a CTL assay of one of the animals. For simplicity, the cytotoxicity for all animals at the same effector/target ratio of 10:1 was deduced from dilution curves, as shown in Fig. 1. The results for all animals, as shown in Table 2, allows a comparison of the breadth of specificity to five different parasite strains exhibited by CTL from the Muguga-immunized and the Muguga cocktail-immunized calves. ANOVA analysis confirmed that there were no statistically difference (P = 0.421). Importantly, CTL from the Muguga-immunized animals killed all five targets to various degrees and no clear strain specificity was observed. CTL from BH047 showed a consistently lower level of killing compared to the other CTL, despite several attempts to establish a bulk culture showing higher cytotoxicity.
Fig. 1.
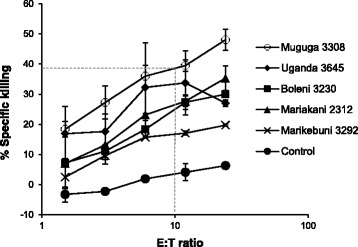
Example of the result from a CTL assay. Serial dilutions of CTL from BG052 were tested for lysis of fixed numbers of the various target cells shown in the figure. The dashed line represents the deduced specific killing at an effector:target ratio (E:T ratio) of 10:1 for the target Muguga 3308. This method was used to compare killing of the various target cells as listed in Table 2. Each point represents the average of a double-determination with the SD shown
Table 2.
Cytotoxic T cell responses in immunized cattle assayed on target cells infected with different cloned T. parva strains
| Muguga | Uganda 3308 |
Mariakani 3645 |
Boleni 3212 |
Marikebuni 3230 |
Control 3292 |
|
|---|---|---|---|---|---|---|
| BG033 (M) | 34 | 13 | 35 | 16 | ND | 0 |
| BG042 (C) | 26 | 18 | 17 | 9 | 18 | 0 |
| BG053 (M) | 59 | 40 | 14 | 20 | 34 | 0 |
| BG051 (C) | 51 | 25 | 38 | 22 | 28 | 3 |
| BG052 (M) | 38 | 33 | 25 | 23 | 17 | 4 |
| BG056 (C) | 3 | 22 | 20 | 11 | 13 | 3 |
| BH055 (M) | 40 | 30 | 26 | 13 | 20 | 0a |
| BH047 (C) | 10 | 8 | 8 | 7 | 4 | 0 |
Cytotoxic T cell responses using autologous PBMC infected with different cloned T. parva strains as target cells. The specific cytotoxicity at effector/target ratios of 10:1 are shown. Each value is deduced from an effector (CTL) titration curve as shown in Fig. 1. (M) Muguga 3308 immunized; (C) Muguga cocktail immunized. (ND) Not determined, (a) Control was autologous PBMC. Relative differences of the CTL specificities, between M and C, were tested using ANOVA analysis (P = 0.421)
CTL from the Muguga cocktail-immunized animals were also assessed for recognition of all three components present in the Muguga cocktail. As seen in Table 3, all Muguga cocktail-immunized animals showed killing of the three components. Interestingly, the Muguga component was killed more effectively in all cases (P < 0.001 for each of the cattle) except BH047, where the killing of the Serengeti component was slightly greater (not statistically significant). As there can be variability in CTL assays, these experiments were done at least twice, and for some animals three times, and the results were similar.
Table 3.
Cytotoxic T cell responses in cattle immunized with the Muguga cocktail on the three T. parva component strains as targets
| Muguga 4230 |
Serengeti 4229 |
Kiambu 5 4228 |
Control | |
|---|---|---|---|---|
| BG042 | 26a | 18 | 13 | 0 |
| BG051 | 40a | 28 | 34 | 0 |
| BG056 | 27a | 22 | 15 | 0 |
| BH047 | 9 | 11 | 5 | 0 |
Cytotoxic T cell responses by vaccine immunized animals using autologous PBMC infected with the three different T. parva component strains as targets. Each CTL was titrated on a fixed number of target cells. The effector/target ratio of 10:1 is shown and each value is deduced from an effector (CTL) titration curve as shown in Fig. 1. Statistical comparison of parameters from fitted curves were used to test differences in cytotoxicity between targets. (a) significant higher than for both other targets for each animal (p < 0.001)
Cloned CTL from a haplotype-matched pair show similar strain specificities
To examine the CTL specificity in more detail, we generated CTL clones from one haploidentical pair, namely the A12/A14 pair of BG053 and BG051. We also successfully generated clones from BG052 (results not shown) but not from the rest of the animals. Each clone was tested for cytotoxicity using the five different cloned sporozoite strains as target cells. Figure 2a shows a heat map of the clonal analysis of BG053 which was immunized with Muguga 3308. It is clear from the pattern of recognition that nine different clonotypes from BG053 were identified, with some clones recognizing only one or two strains, and others recognizing three or four of the five strains. The least recognized strain was Marikebuni, and only one clone was found that recognized Muguga alone. In general, the clonal analysis corresponded well with the bulk analysis, where strongest killing was also observed with the Muguga and Uganda stabilates. On the other hand, few of the clones recognized Marikebuni in contrast to the results observed with the bulk cell lines.
Fig. 2.
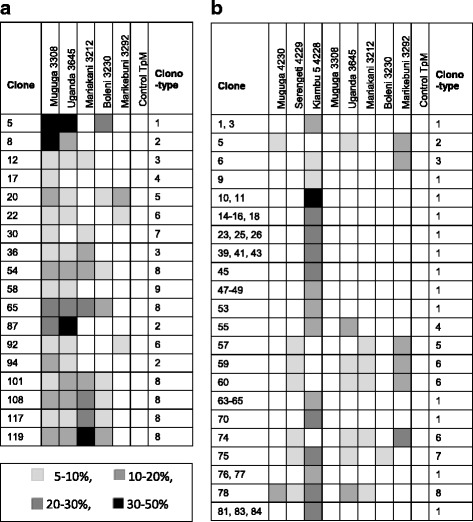
Cytotoxicity obtained by T cell clones on a panel of target cells infected with different cloned T. parva strains. The cutoff value was 5% cytotoxicity. Clones were categorized into clonotypes based on their pattern of reactivity. The level of cytotoxicity is visualized as a heat map – colour codes are shown. a Calf BG053 immunized with Muguga 3308. CD8 T cell clones were generated from CD8-purified bulk cultures and tested for cytotoxicity to 5 different strains (as shown) and a MHC-mismatched control TpM. b The haplotype-matched calf BG051 immunized with the Muguga cocktail 0801. Clones were tested for cytotoxicity on the same target cells as BG053 and the additional components of the Muguga Cocktail
The clonal analysis of the Muguga cocktail-immunized animal BG051 is shown in Fig. 2b. Eight clonotypes were identified, with a surprisingly high number of clones specific for Kiambu5 only. There were also clones which recognized a broader set of targets. As observed in the analysis of bulk CTL lines, there was no evidence from the clonal analysis of a wider set of clonal reactivities in the Muguga cocktail-immunized animal compared to the Muguga only-immunized animal. Table 4 shows the number of clones with the percentages in brackets, that recognize indicated numbers of different cloned strains (upper part), and the number of clones with percentage in brackets recognizing a particular parasite strain (lower part). It is evident that a large fraction of clones from the Muguga cocktail-immunized animal recognized one strain only, whereas more clones from the Muguga only-immunized animal recognized many of the strains, which is opposite of what was expected. All clones from the Muguga only-immunized animal recognized Muguga as expected, and 89% recognized Uganda, implying that this strain is quite similar to Muguga. On the other hand, Marikebuni was the least recognized strain, being specifically lysed by only 17% of the clones. In the Muguga cocktail-immunized animal, 87% of the clones recognized Kiambu5, one of the components in the vaccine, compared to 5% and 16% that recognized the Muguga and Serengeti-transformed components, respectively. Interestingly, there was a difference in the two Muguga-infected targets, which may reflect qualitative differences in presented epitopes in these two target cells.
Table 4.
Number and percentages of CTL clones from BH053 and BH051 recognizing multiple T. parva strains
| Calf | BG053 (M) | BG051 (C) |
|---|---|---|
| Total | 18 | 38 |
| 1 strain | 1 (6) | 29 (76) |
| 2 strains | 5 (28) | 2 (5) |
| 3 strains | 5 (28) | 2 (5) |
| 4 strains | 7 (39) | 4 (11) |
| 5 strains | 0 (0) | 1 (3) |
| Muguga (4230) | ND | 2 (5) |
| Serengeti (4229) | ND | 6 (16) |
| Kiambu-5 (4228) | ND | 33 (87) |
| Muguga (3308) | 18 (100) | 2 (5) |
| Uganda (3645) | 16 (89) | 7 (18) |
| Mariakani (3212) | 9 (50) | 5 (13) |
| Boleni (3230) | 8 (44) | 1 (3) |
| Marikebuni (3292) | 3 (17) | 6 (16) |
Upper part of the table: The total number of clones with percentages in brackets, that recognize multiple cloned T. parva strains, with a cutoff value of 5% cytotoxicity, for the haplotype matched pair, BG053 (Muguga immunized) and BG051 (Muguga cocktail immunized), is shown. Each clone was analyzed for the number of strains that it recognized. The left numbers in each column represents the actual number of clones recognizing 1 strain, 2 strains, etc. The numbers in brackets represents the corresponding percentages of clones recognizing 1 strain, 2 strains etc. Percentages have been rounded. Lower part of the table: The total number of clones and percentages in brackets from BG053 and BG051 recognizing the different T. parva strains. Percentages have been rounded. BH051 was tested on the Muguga cocktail component reference stabilate strains in addition to the cloned strains. (ND) Not determined
In summary, the results of the CTL assays do not support the hypothesis that immunization with the Muguga cocktail induces CTL with a broader reactivity against T. parva-infected cell lines compared to immunization with the Muguga stabilate alone, at least with the parasite strains and MHC haplotypes of the animals assessed here.
Similar protection to the vaccine strains in animals vaccinated with Muguga or the Muguga cocktail
The animals were challenged with the Muguga cocktail in order to investigate if there were any differences between the Muguga-immunized animals and the Muguga cocktail-immunized animals in their immunity to parasite strains present in the Muguga cocktail vaccine but not in the Muguga stabilate. Two non-immunized control animals were used to confirm a sufficient challenge had been delivered and the clinical outcome was assessed with the Rowlands ECF index [30]. The experiment was stopped at day 14 as there was no development of disease in the animals except for the two control animals. As seen in Table 5, there was no substantive difference in the protection to the Muguga cocktail between animals immunized with Muguga only or the Muguga cocktail. Only one animal from each immunized group developed pyrexia, and schizonts were detected in three of the seven animals, two from the Muguga-immunized group and one from the Muguga cocktail-immunized group. This contrasts strongly with the control animals, both of which developed pyrexia and had detectable schizonts and piroplasms. Interestingly, the animal with the highest ECF score, BH047, was the animal with the weakest CTL response.
Table 5.
Challenge of cattle with the Muguga cocktail 0801
| Animal | Immunization | Days with Schizonts | Days with pyrexia | Days with piroplasms | Day treated | ECF index |
|---|---|---|---|---|---|---|
| BG033 | Mug 3308 (M) | 5 | 1.04 | |||
| BG053 | Mug 3308 (M) | 2 | 2.21 | |||
| BH055 | Mug 3308 (M) | 2 | 2.43 | |||
| BG051 | Vac 0801 (C) | 1.04 | ||||
| BG042 | Vac 0801 (C) | 1.04 | ||||
| BH047 | Vac 0801 (C) | 6 | 4 | 2.98 | ||
| BG056 | Vac 0801 (C) | 1.04 | ||||
| BG039 | None | 7 | 5 | 2 | 13 | 6 |
| BH043 | None | 7 | 4 | 2 | 13 | 5.93 |
All 7 immunized cattle (BG052 succumbed for unknown reasons) and two naïve control cattle BG039, BH043 were challenged with the Muguga cocktail vaccine stabilate 0801. Number of days with schizonts, pyrexia, piroplasms and the days until treatment are indicated. The ECF index is also indicated
Discussion
It is of major importance to fully understand the mechanisms underlying the protective immune response to T. parva, both to underpin the deployment of the current live Muguga cocktail vaccine and to guide the development of a subunit vaccine.
The Muguga cocktail vaccine is composed of parasites from three different isolates (Muguga, Kiambu 5 and Serengeti-transformed), and it is known to protect animals efficiently when deployed in the field [5], despite the presence of antigenic variation in field populations of the parasite [31]. The experiments reported in this article were undertaken to demonstrate that the trivalent Muguga cocktail vaccine provided a more complete protection against heterologous challenge than the Muguga stabilate alone by inducing CTLs capable of recognizing a broader range of parasite strains.
Surprisingly, there was no substantive difference in the CTL responses to the different T. parva strains between MHC-matched animals immunized with the Muguga cocktail and the Muguga only. The clonal analysis of one of the haplotype matched pairs supported these results (unfortunately we were not able to generate clones from the other haplotype matched pairs). We were surprised to see the many Kiambu 5-specific clones from the vaccine immunized BG051 animal. This result did not corroborate with the bulk result (Table 2), where there were good responses to Muguga and to Serengeti. It is possible that there is a bias in the cloning process in some cases. Some clones could be easier to expand than others, which would limit the interpretation of the clonal analyses in general. Nevertheless, the bulk results also did not favour a broader CTL specificity in the Muguga cocktail immunized animals compared to those immunized with the Muguga stabilate. An interesting observation was that certain clones recognized many different target cells, which shows that there are indeed broadly cross-reactive antigenic determinants which induce a CTL response following ITM immunization, either with Muguga alone or with the Muguga cocktail. Future work will aim to map these epitopes, which could be useful in a subunit vaccine.
Previous research on the strain specificity of the CTL response in animals immune to T. parva suggests that the response in each animal is dominated by a small number of antigens [10, 11, 19, 20]. Clonal analysis of the response in MHC-homozygous animals confirmed this immunodominance by showing that over 60% of the clones from the animals recognized single epitopes in the two respective antigens presented by the MHC haplotypes [24]. The relatively high number of clonotypes observed in the A12/A14 animals analysed here suggests that this is not the case in these animals, unless there is an immunodominant antigen which displays antigenic diversity. In other words, the differential reactivity of the clonotypes is a consequence of the different levels of cross-reactivity of each clone with variant forms of a dominant epitope. The T. parva antigens recognized by the various MHC alleles in the A12 and A14 animals have not been fully identified so it is currently not possible to assess the recognition of specific epitopes and the level of diversity displayed by such epitopes. The focus of the response in the Muguga cocktail-immunized animal on Kiambu 5 is interesting and may reflect an immunodominant antigen found predominantly in Kiambu and not shared with many other strains. In this respect, it would be interesting to assess whether CTL from Muguga-only immunized animals are cytotoxic towards the three components in the Muguga cocktail, and to assess whether CTL derived from animals immunized with any one of the components comprising the Muguga cocktail recognize the cloned strains as this may reveal if there are immunodominant, cross-reactive epitopes in the stabilates, and whether this outcome is dependent on the MHC background of the animals.
In order to show if the lack of differences in the CTL responses was reflected in vivo, all animals were challenged with a lethal dose of the Muguga cocktail. There were no major differences in the protection observed in the animals, which indicates that there are no important differences in the antigenic composition of the Muguga stabilate and the Muguga cocktail, at least in animals of the MHC types studied here. This result is perhaps not surprising, as the orginal studies of Radley et al., (1975a) showed that the Muguga stabilate provided good protection against both the Kiambu 5 and Serengeti transformed stabilates, and vice versa. In addition, recent deep-sequencing results from our group have also shown that the Muguga cocktail does not contain a great amount of diversity in the known CTL antigens which were examined [32]. Five of the nine antigen genes sequenced were present as a single version, with three present in two forms and the final as three variants. These results suggest that the three components are antigenically very similar and they invoke the question of why the Muguga cocktail provided better protection in the original experiments and why the Muguga cocktail has been so successful in protecting animals against field challenge, where heterogeneous challenges will be far more predominant. It may be, as argued elsewhere [33], that antigenic diversity in the vaccine stabilate is not as essential as originally believed. A note of caution is that most of the antigens which have been sequenced are those presented by predominantly European cattle, and may not reflect important antigens recognized in breeds where the vaccine has been deployed.
These results are somewhat contradictory to the earlier study, where it was shown that a mixture of Muguga, Kiambu5 and Serengeti transformed stabilates provided better protection than individual stabilates [34]. It should be noted that a different challenge strain (Kiambu 1) was used in the earlier experiment. Thus, a possible explanation for the difference in results is that the CTL induced by the Muguga isolate do not cross-react with antigens present in Kiambu 1, at least in the animals of the MHC types used in the earlier experiment. This experiment was performed before the role of CTL as the mediators of immunity and the influence of the MHC were established, and the MHC types of the animals are not available.
However, the difference in the results does indicate that, although we have shown that Muguga-immunized animals of the MHC types used here can generate CTL of similar strain specificities as those immunized with the Muguga cocktail, we cannot with certainty state that use of the Muguga isolate alone will provide the same broad protection in the field as the Muguga cocktail appears to provide. In this respect, it would also be interesting to test if cattle immunized with either Muguga only or the Muguga cocktail would be protected if challenged with the strains, that previously were shown to break through single-strain immunization, such as Marikebuni and Kiambu 1, this time with a larger number of cattle per group.
The field situation is far more complicated than the experimental conditions employed here, due to the presence of a much more heterogenous population of parasites and the diversity of MHC types in outbred cattle populations of several breeds. Both of these factors threaten the success of vaccines composed of a limited number of parasite strains. Particularly at risk are cattle populations in the buffalo-cattle interface, as parasites derived from buffalo show much greater antigenic diversity than those from cattle [31]. Indeed, it has been shown that immunization with the Muguga cocktail does not protect cattle in areas of close interaction with buffalo [35], although it has not been established that this is due to antigenic diversity. Close monitoring of break-through incidences in the field with use of the Muguga cocktail versus single strains would show whether or not single strains protect as well as the Mugaga cocktail.
Conclusion
There were no indications that a broader immune response was induced by immunization with the Muguga cocktail compared to the Muguga strain only, in the haplotypes examined in this study. In agreement with this, previous studies on antigenic diversity in the Muguga cocktail found limited diversity. As the original studies using single strain and Muguga cocktail for induction of protection were performed with limited number of animals and with high doses of needle challenges, this may warrant for testing single vaccine strains in field settings where the load of parasites during challenge will be much lower.
Acknowledgements
We wish to thank Stephen Mwaura for assisting with the immunization and monitoring of the cattle and Jane Poole, Research and Methods Group, ILRI, for performing the statistical analyses.
Funding
This work was partially supported by a grant awarded jointly by the Department for International Development (UK Government) and the Biotechnology and Biological Sciences Research Council (BBSRC) UK grant number BB/H009515/1 of the Combating Infectious Diseases of Livestock for International Development (CIDLID) program. Additional support was received from the CGIAR Research Program on Livestock and Fish, led by ILRI. We also acknowledge the CGIAR Fund Donors (http://www.cgiar.org/funders).
Availability of data and materials
The datasets generated and/or analysed during the current study are available from the corresponding author on reasonable request.
Abbreviations
- ConA
Concanavalin A
- CTL
Cytotoxic T Lymphocytes
- FBS
Fetal Bovine Serum
- IFNγ
Interferon Gamma
- ITM
Infection and Treatment Method
- mAb
Monoclonal Antibody
- MHC
Major Histocompatibility Complex
- PBMC
Peripheral Blood Mononuclear Cells
- SD
Standard Deviation
- TCGF
T-Cell Growth Factor
- TCR
T Cell Receptor
Authors’ contributions
LS designed the study, interpreted results and wrote the manuscript. NS performed the MHC Class I typing, EA performed the CTL assay, TN sourced animals and did the clinical laboratory assessments, RS was involved in generation of cell lines and CTL assays. IM was involved in the overall conceptualization, PT was involved in the design, result interpretation and helped with writing of the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The animal study was approved by the ILRI Institutional Animal Care and Use Committee, IACUC No: 2010.07. Animal owner consents were not necessary, as all animals were bought and brought to the ILRI farm, before experiments were performed.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Lucilla Steinaa, Phone: +254 20 422 3379, Email: l.steinaa@cgiar.org.
Nicholas Svitek, Email: n.svitek@cgiar.org.
Elias Awino, Email: e.awino@cgiar.org.
Thomas Njoroge, Email: t.njoroge@cgiar.org.
Rosemary Saya, Email: r.saya@cgiar.org.
Ivan Morrison, Email: ivan.morrison@roslin.ed.ac.uk.
Philip Toye, Email: p.toye@cgiar.org.
References
- 1.Hulliger L, Wilde KH, Brown CG, Turner L. Mode of multiplication of Theileria in cultures of bovine lymphocytic cells. Nature. 1964;203:728–730. doi: 10.1038/203728a0. [DOI] [PubMed] [Google Scholar]
- 2.McKeever DJ, Morrison WI. Immunity to a parasite that transforms T lymphocytes. Curr Opin Immunol. 1994;6:564–567. doi: 10.1016/0952-7915(94)90142-2. [DOI] [PubMed] [Google Scholar]
- 3.Morrison WI, Goddeeris BM, Teale AJ, Groocock CM, Kemp SJ, Stagg DA. Cytotoxic T-cells elicited in cattle challenged with Theileria parva (Muguga): evidence for restriction by class I MHC determinants and parasite strain specificity. Parasite Immunol. 1987;9:563–578. doi: 10.1111/j.1365-3024.1987.tb00530.x. [DOI] [PubMed] [Google Scholar]
- 4.Radley DE, Brown CGD, Cunningham MP. East Coast fever: 1. Chemprophylactic immunization of cattle against Theileria parva (Muguga) and five Thelerial strains. Vet Parasitol. 1975;1:35–41. doi: 10.1016/0304-4017(75)90005-9. [DOI] [Google Scholar]
- 5.Di Giulio G, Lynen G, Morzaria S, Oura C, Bishop R. Live immunization against East Coast fever--current status. Trends Parasitol. 2009;25:85–92. doi: 10.1016/j.pt.2008.11.007. [DOI] [PubMed] [Google Scholar]
- 6.Martins SB, Di Giulio G, Lynen G, Peters A, Rushton J. Assessing the impact of East Coast fever immunisation by the infection and treatment method in Tanzanian pastoralist systems. Prev Vet Med. 2010;97:175–182. doi: 10.1016/j.prevetmed.2010.09.018. [DOI] [PubMed] [Google Scholar]
- 7.Irvin AD, Mwamachi DM. Clinical and diagnostic features of East Coast fever (Theileria parva) infection of cattle. Vet Rec. 1983;113:192–198. doi: 10.1136/vr.113.9.192. [DOI] [PubMed] [Google Scholar]
- 8.Irvin AD, Dobbelaere DA, Mwamachi DM, Minami T, Spooner PR, Ocama JG. Immunisation against East Coast fever: correlation between monoclonal antibody profiles of Theileria parva stocks and cross immunity in vivo. Res Vet Sci. 1983;35:341–346. [PubMed] [Google Scholar]
- 9.Emery DL, Eugui EM, Nelson RT, Tenywa T. Cell-mediated immune responses to Theileria parva (East Coast fever) during immunization and lethal infections in cattle. Immunology. 1981;43:323–336. [PMC free article] [PubMed] [Google Scholar]
- 10.Goddeeris BM, Morrison WI, Teale AJ. Generation of bovine cytotoxic cell lines, specific for cells infected with the protozoan parasite Theileria parva and restricted by products of the major histocompatibility complex. Eur J Immunol. 1986;16:1243–1249. doi: 10.1002/eji.1830161010. [DOI] [PubMed] [Google Scholar]
- 11.Goddeeris BM, Morrison WI, Teale AJ, Bensaid A, Baldwin CL. Bovine cytotoxic T-cell clones specific for cells infected with the protozoan parasite Theileria parva: parasite strain specificity and class I major histocompatibility complex restriction. Proc Natl Acad Sci U S A. 1986;83:5238–5242. doi: 10.1073/pnas.83.14.5238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Allen TM, Sidney J, del Guercio MF, Glickman RL, Lensmeyer GL, Wiebe DA, DeMars R, Pauza CD, Johnson RP, Sette A, Watkins DI. Characterization of the peptide binding motif of a rhesus MHC class I molecule (Mamu-a*01) that binds an immunodominant CTL epitope from simian immunodeficiency virus. J Immunol. 1998;160:6062–6071. [PubMed] [Google Scholar]
- 13.Altfeld MA, Trocha A, Eldridge RL, Rosenberg ES, Phillips MN, Addo MM, Sekaly RP, Kalams SA, Burchett SA, McIntosh K, et al. Identification of dominant optimal HLA-B60- and HLA-B61-restricted cytotoxic T-lymphocyte (CTL) epitopes: rapid characterization of CTL responses by enzyme-linked immunospot assay. J Virol. 2000;74:8541–8549. doi: 10.1128/JVI.74.18.8541-8549.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McMurtrey CP, Lelic A, Piazza P, Chakrabarti AK, Yablonsky EJ, Wahl A, Bardet W, Eckerd A, Cook RL, Hess R, et al. Epitope discovery in West Nile virus infection: identification and immune recognition of viral epitopes. Proc Natl Acad Sci U S A. 2008;105:2981–2986. doi: 10.1073/pnas.0711874105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Provenzano M, Mocellin S, Bettinotti M, Preuss J, Monsurro V, Marincola FM, Stroncek D. Identification of immune dominant cytomegalovirus epitopes using quantitative real-time polymerase chain reactions to measure interferon-gamma production by peptide-stimulated peripheral blood mononuclear cells. J Immunother. 2002;25:342–351. doi: 10.1097/00002371-200207000-00006. [DOI] [PubMed] [Google Scholar]
- 16.Bakker AB, Schreurs MW, Tafazzul G, de Boer AJ, Kawakami Y, Adema GJ, Figdor CG. Identification of a novel peptide derived from the melanocyte-specific gp100 antigen as the dominant epitope recognized by an HLA-A2.1-restricted anti-melanoma CTL line. Int J Cancer. 1995;62:97–102. doi: 10.1002/ijc.2910620118. [DOI] [PubMed] [Google Scholar]
- 17.Ghosh A, Wolenski M, Klein C, Welte K, Blazar BR, Sauer MG. Cytotoxic T cells reactive to an immunodominant leukemia-associated antigen can be specifically primed and expanded by combining a specific priming step with nonspecific large-scale expansion. J Immunother. 2008;31:121–131. doi: 10.1097/CJI.0b013e31815aaf24. [DOI] [PubMed] [Google Scholar]
- 18.Tzelepis F, de Alencar BC, Penido ML, Claser C, Machado AV, Bruna-Romero O, Gazzinelli RT, Rodrigues MM. Infection with Trypanosoma cruzi restricts the repertoire of parasite-specific CD8+ T cells leading to immunodominance. J Immunol. 2008;180:1737–1748. doi: 10.4049/jimmunol.180.3.1737. [DOI] [PubMed] [Google Scholar]
- 19.Taracha EL, Goddeeris BM, Morzaria SP, Morrison WI. Parasite strain specificity of precursor cytotoxic T cells in individual animals correlates with cross-protection in cattle challenged with Theileria parva. Infect Immun. 1995;63:1258–1262. doi: 10.1128/iai.63.4.1258-1262.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Taracha EL, Goddeeris BM, Teale AJ, Kemp SJ, Morrison WI. Parasite strain specificity of bovine cytotoxic T cell responses to Theileria parva is determined primarily by immunodominance. J Immunol. 1995;155:4854–4860. [PubMed] [Google Scholar]
- 21.Bishop R, Nene V, Staeyert J, Rowlands J, Nyanjui J, Osaso J, Morzaria S, Musoke A. Immunity to East Coast fever in cattle induced by a polypeptide fragment of the major surface coat protein of Theileria parva sporozoites. Vaccine. 2003;21:1205–1212. doi: 10.1016/S0264-410X(02)00621-7. [DOI] [PubMed] [Google Scholar]
- 22.Gardner MJ, Bishop R, Shah T, de Villiers EP, Carlton JM, Hall N, Ren Q, Paulsen IT, Pain A, Berriman M, et al. Genome sequence of Theileria parva, a bovine pathogen that transforms lymphocytes. Science. 2005;309:134–137. doi: 10.1126/science.1110439. [DOI] [PubMed] [Google Scholar]
- 23.Graham SP, Pelle R, Honda Y, Mwangi DM, Tonukari NJ, Yamage M, Glew EJ, de Villiers EP, Shah T, Bishop R, et al. Theileria parva candidate vaccine antigens recognized by immune bovine cytotoxic T lymphocytes. Proc Natl Acad Sci U S A. 2006;103:3286–3291. doi: 10.1073/pnas.0511273103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.MacHugh ND, Connelley T, Graham SP, Pelle R, Formisano P, Taracha EL, Ellis SA, McKeever DJ, Burrells A, Morrison WI. CD8+ T-cell responses to Theileria parva are preferentially directed to a single dominant antigen: implications for parasite strain-specific immunity. Eur J Immunol. 2009;39:2459–2469. doi: 10.1002/eji.200939227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ellis SA, Staines KA, Stear MJ, Hensen EJ, Morrison WI. DNA typing for BoLA class I using sequence-specific primers (PCR-SSP) Eur J Immunogenet. 1998;25:365–370. doi: 10.1046/j.1365-2370.1998.00112.x. [DOI] [PubMed] [Google Scholar]
- 26.Patel EH, Lubembe DM, Gachanja J, Mwaura S, Spooner P, Toye P. Molecular characterization of live Theileria parva sporozoite vaccine stabilates reveals extensive genotypic diversity. Vet Parasitol. 2011;179:62–68. doi: 10.1016/j.vetpar.2011.01.057. [DOI] [PubMed] [Google Scholar]
- 27.Goddeeris BM, Morrison WI. Techniques for generation, cloning, and charachterization of bovine cytotoxic T cells specific for the protozoan Theileria parva. J Tiss Culture Methods. 1988;11:101. doi: 10.1007/BF01404140. [DOI] [Google Scholar]
- 28.Morzaria SP, Dolan TT, Norval RA, Bishop RP, Spooner PR. Generation and characterization of cloned Theileria parva parasites. Parasitology. 1995;111(Pt 1):39–49. doi: 10.1017/S0031182000064581. [DOI] [PubMed] [Google Scholar]
- 29.Svitek N, Taracha EL, Saya R, Awino E, Nene V, Steinaa L. Analysis of the cellular immune responses to vaccines. Methods Mol Biol. 2016;1349:247–262. doi: 10.1007/978-1-4939-3008-1_16. [DOI] [PubMed] [Google Scholar]
- 30.Rowlands GJ, Musoke AJ, Morzaria SP, Nagda SM, Ballingall KT, McKeever DJ. A statistically derived index for classifying East Coast fever reactions in cattle challenged with Theileria parva under experimental conditions. Parasitology. 2000;120(Pt 4):371–381. doi: 10.1017/S0031182099005600. [DOI] [PubMed] [Google Scholar]
- 31.Pelle R, Graham SP, Njahira MN, Osaso J, Saya RM, Odongo DO, Toye PG, Spooner PR, Musoke AJ, Mwangi DM, et al. Two Theileria parva CD8 T cell antigen genes are more variable in buffalo than cattle parasites, but differ in pattern of sequence diversity. PLoS One. 2011;6:e19015. doi: 10.1371/journal.pone.0019015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hemmink JD, Weir W, MacHugh ND, Graham SP, Patel E, Paxton E, Shiels B, Toye PG, Morrison WI, Pelle R. Limited genetic and antigenic diversity within parasite isolates used in a live vaccine against Theileria parva. Int J Parasitol. 2016;46:495–506. doi: 10.1016/j.ijpara.2016.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Morrison WI, Connelley T, Hemmink JD, MacHugh ND. Understanding the basis of parasite strain-restricted immunity to Theileria parva. Annu Rev Anim Biosci. 2015;3:397–418. doi: 10.1146/annurev-animal-022513-114152. [DOI] [PubMed] [Google Scholar]
- 34.Radley DE, Brown CG, Cunningham MP, Kimber CD, Musisi FL, Payne RC, Purnell RE, Stagg DA, Young AS. East cost fever:3. Chemoprophylactic immunization of cattle using oxytetracycline and a combination of Theileria strains. Vet Parasitol. 1975;1:51–60. doi: 10.1016/0304-4017(75)90007-2. [DOI] [Google Scholar]
- 35.Sitt T, Poole EJ, Ndambuki G, Mwaura S, Njoroge T, Omondi GP, Mutinda M, Mathenge J, Prettejohn G, Morrison WI, Toye P. Exposure of vaccinated and naive cattle to natural challenge from buffalo-derived Theileria parva. Int J Parasitol Parasites Wildl. 2015;4:244–251. doi: 10.1016/j.ijppaw.2015.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets generated and/or analysed during the current study are available from the corresponding author on reasonable request.


