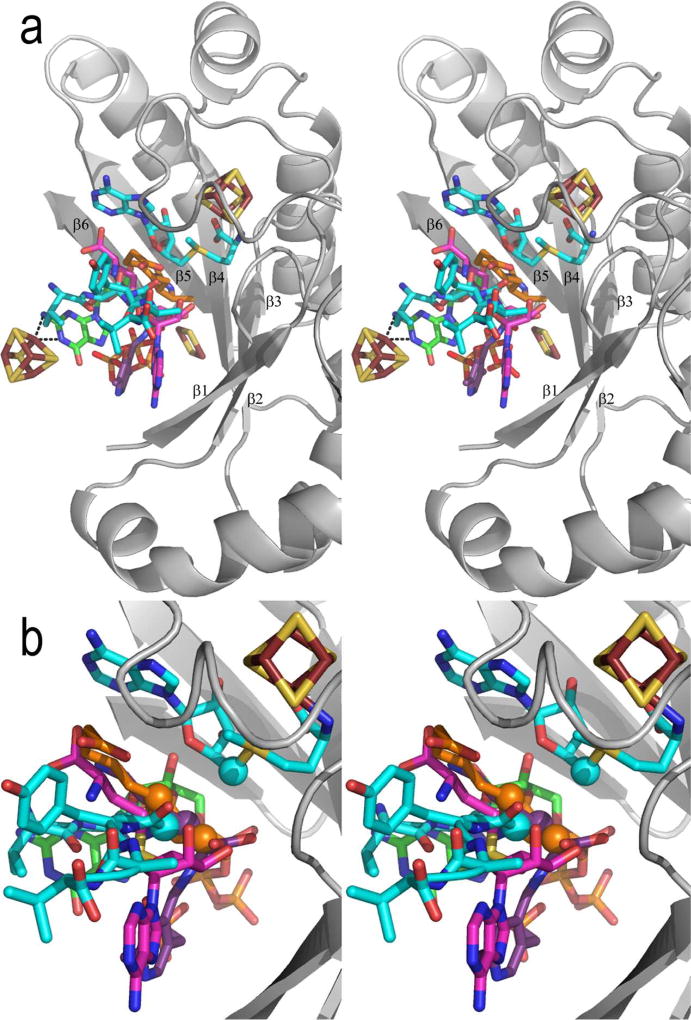
Figure 8. Stereoview of the substrate binding sites of the Radical SAM enzymes.
In (a), five structures with substrate bound are superimposed based on their Radical SAM cores. Shown is the Radical SAM core, 4Fe-4S cluster and AdoMet of PFL-AE only, with AdoMet carbons colored teal. The five “substrates” of the enzymes are shown in sticks, colored as follows: teal, PFL-AE; magenta, HemN; orange, BioB; green, MoaA; purple, LAM. PLP of LAM and the BioB 2Fe-2S, MoaA 4Fe-4S and HydE 2Fe-2S clusters are also shown, displayed as in other figures. In (b), five enzyme structures are superimposed on their 4Fe-4S clusters and AdoMet only, to give a more accurate comparison of the relative positions of the substrates with respect to AdoMet. The core backbone, 4Fe-4S cluster and AdoMet of PFL-AE are shown, and colors are as described in (a). C5' of AdoMet is shown as a sphere. The atoms from which hydrogen abstraction is known to occur (i.e. Cα of G734 of the peptide, C6 and C8 of dethiobiotin, and Cβ of lysine) are also shown as spheres.