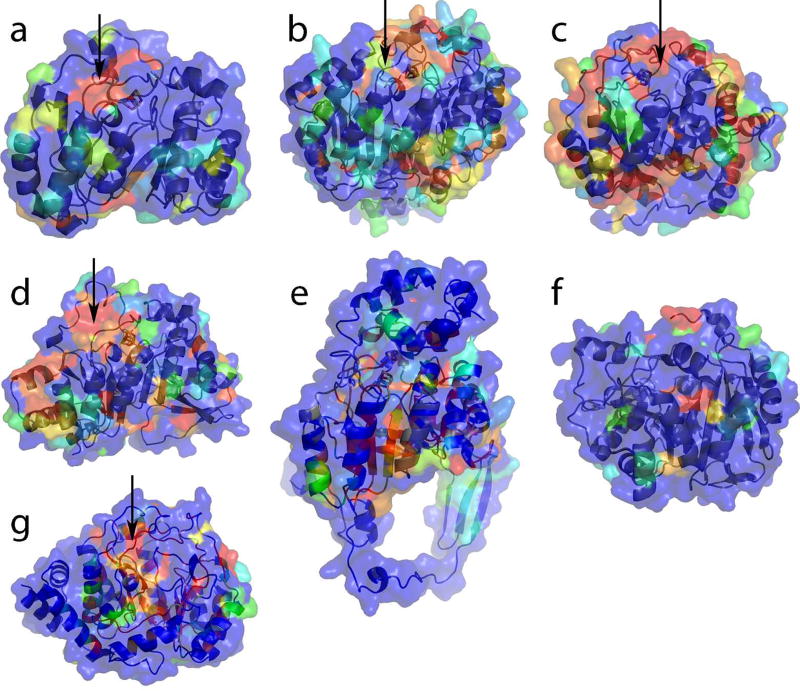
Figure 9. Surface conservation of Radical SAM enzymes suggests putative binding site for physiological reductant.
Shown are the cartoon representations and transparent surfaces of (a) PFL-AE, (b) HemN, (c) BioB, (d) MoaA, (e) LAM, (f) phTYW1 and (g) HydE, from the the opposite side of the partial barrel with respect to the lateral opening of the Radical SAM core, in roughly the same view as in Figure 3b. The transparent surface is colored as a rainbow according to the extent of sequence conservation with respect to the protein’s individual family, with red being 100% conserved and blue as 0% conserved. A BLAST search was conducted via the ExPASy proteomics server using the sequence of the enzyme that is structurally characterized as the query. The top matching sequences (up to 100 in number) were input for alignment by ClustalW. The alignment output by ClustalW was then used as input to ESPript, which calculated the extent of conservation of each residue. Arrows indicate proposed surface for interaction with reductant. See Figure S16 for stereoviews of each surface.