Fig. 1.
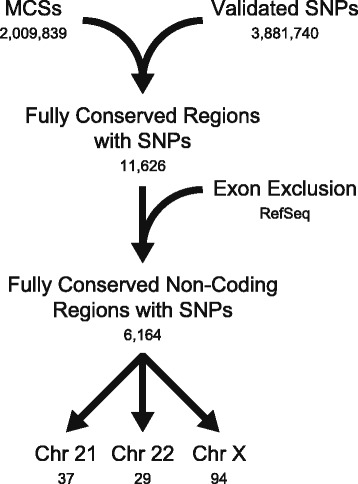
A computational pipeline to identify putative regulatory SNPs. The human, mouse, and chicken genomes were aligned, and genomic segments five base-pairs in length or greater and identical in all three species were identified to compile a panel of multiple-species conserved sequences (MCSs). Overlap between the MCS dataset and validated ‘by-frequency’ SNPs from db SNP130 was determined. Exons were excluded using RefSeq entries, and a pilot set of 160 regions were identified on chromosomes 21, 22, and X. The number of regions in each resulting dataset are indicated below the label
