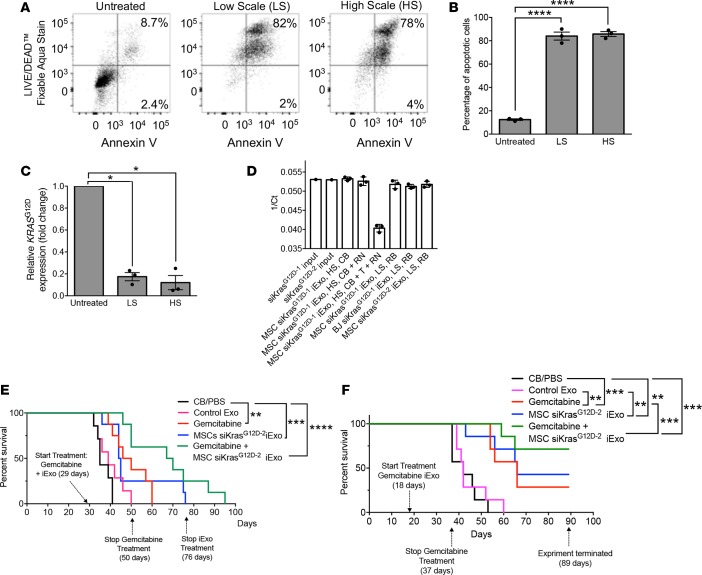
Figure 6. Efficacy of large-scale produced-GMP iExosomes in combination with gemcitabine.
(A) Representative dot plot and (B) quantification of flow cytometry analyses of apoptosis in Panc-1 cells induced by MSCs siKrasG12D–2 iExo, comparing low scale (LS) or high scale (HS) electroporation of MSC exosomes. Numbers represent the percentage of positive cells (n = 3 independent experiments, 1-way ANOVA compared with untreated). (C) KRASG12D transcript levels in Panc-1 cells incubated 3 hours with MSCs siKrasG12D–2, comparing LS or HS electroporation of MSC exosomes (n = 3 independent experiments, 1-tailed unpaired t test). (D) qPCR of siRNA for KrasG12D (same siRNA sequence from 2 purchasing sources, siKrasG12D–1 and siKrasG12D–2 for source 1 and 2, respectively) in the indicated samples (n = 3 distinct samples treated on the same day; input siRNA: n = 1). The data are presented as 1/Ct and mean ± SD. CB, clinical buffer; RB, research buffer; T, Triton X-100; RN, RNase A. (E) Kaplan-Meier curve indicating the survival of KPC689 mice after tumor induction in the listed treatment groups (CB/PBS [n = 7], Control Exo [n = 7], gemcitabine [n = 8], MSC siKrasG12D–2 iExo [n = 8], gemcitabine + MSCs siKrasG12D–2 iExo [n = 8]; log-rank [Mantel-Cox] test). (F) Kaplan-Meier curve indicating the survival of KPC689 mice after tumor induction in the listed treatment groups (n = 7 mice in each of the listed groups; log-rank [Mantel-Cox] test). Unless otherwise specified, mean ± SEM is depicted. Unless stated otherwise, 1-way ANOVA, comparing experimental groups to control groups, was used to determine statistical significance. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. See Supplemental Source Data 1 and 2.