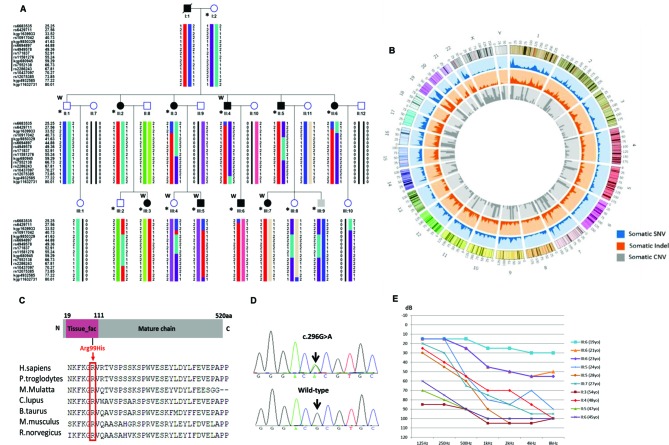
Figure 1.
Identification of a mutation in IFNLR1 causing ADNSHL. (A) The pedigree of a three-generational family with segregating progressive ADNSHL. DNA samples were obtained from six unaffected and nine affected individuals (denoted by asterisks). Individuals who underwent whole-exome sequencing and linkage analysis are marked with W. Black symbols represent affected individuals. The haplotype created with SNP markers shows complete cosegregation of a locus at 1p34.1–1 p36.12 (red) in family JS4842. (B) Distribution of variants in the chromosome. Blue represents the distribution of somatic SNVs, orange indicates the distribution of somatic Indel mutations and grey represents the distribution of somatic CNVs. (C) Schematic diagram of IFNLR1 and protein alignment showing IFNLR1 Arg99His occurred at evolutionarily conserved amino acids (in red box) within the tissue_fac domain across nine species. (D) Electropherograms showing that the IFNLR1 c.296G>A mutation cosegregated with hearing loss. (E) Audiograms from selected individuals of family JS4842. Thresholds reveal the presence of moderate to severe sensorineural hearing loss, which occurred at a late onset in all affected individuals. For clarity, thresholds are shown for one ear from each individual. No differences in hearing were detected when the two ears were compared in an affected individual. Younger individuals had less severe hearing loss compared with the older affected individuals, suggesting that hearing loss in this family is progressive in nature. ADNSHL, autosomal-dominant, non-syndromic, progressive sensorineural hearing loss; IFNLR1, interferon lambda receptor 1; SNV, single nucleotide variant.