Figure 6. Association of predicted sequence-paired sites and monomeric Notch response elements with dimer-dependent and -independent Notch target genes in human T-ALL cells.
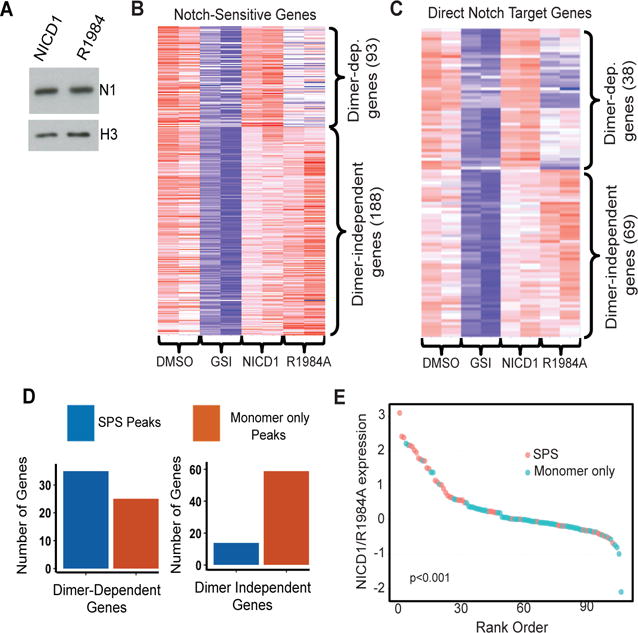
(A) Abundance of wild type NICD1 and R1984A mutant NICD1 in transduced CUTLL1 cells. (B and C) Heat maps of Notch-dependent RNA expression (B) and high confidence Notch target gene expression (C) in cells treated with vehicle (DMSO), GSI (1μM DBZ), or GSI following transduction with wild type or R1984A mutant NICD1. Expression was determined by sequencing RNA obtained from two independent experiments. (D) Notch dimer-dependent and –independent genes are associated with predicted sequence-paired sites and monomeric RBPJ/NOTCH1 sites, respectively. Notch regulated genes were paired with nearby predicted SPS or monomeric RBPJ/NOTCH1 binding sites as described (17). Genes rescued by R1984A expression in the presence of GSI are preferentially associated with monomeric RBPJ/NOTCH1 binding sites than with SPSs (p < 1 × 10−5, Fisher Exact Test). (E) Waterfall plot showing the association between the dimer-dependency of likely direct Notch target genes and the presence of one or more nearby SPSs (p<1 × 10−3, Wilcoxon one-sided signed-rank test). The Y-axis corresponds to the log2 expression ratio of individual Notch target genes in GSI treated cells transduced with wild type or R1984A mutant NICD1.
