Figure 7. Co-regulation of HES5 and HANR1 by enhancer RBPJ/NOTCH1 binding sites.
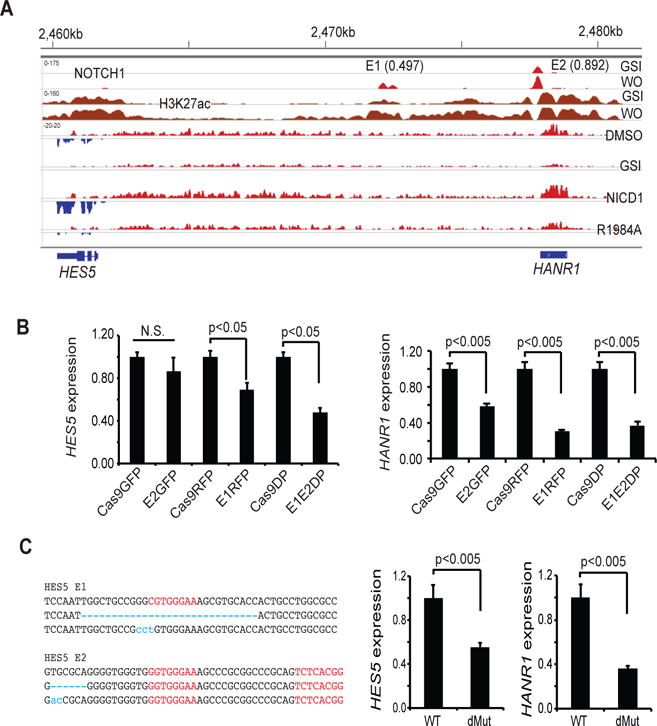
(A) HES5/HANR1 co-regulation. Upper panels show NOTCH1 and H3K27ac aligned read from ChIP-Seq data for CUTLL1 cells in the Notch-off (GSI) and Notch-on (GSI washout [WO]). Dynamic NOTCH1 binding sites designated E1 and E2 have PBM product scores of 0.497 and 0.892, respectively. Lower panels show stranded RNA-seq data obtained from CUTLL1 cells treated with vehicle, GSI (1μM DBZ), or DBZ following transducing of wild type NICD1 or the R1984A NICD1 mutant. HANR1 and HES5 aligned reads from opposite DNA strands are shown in red and blue, respectively. (B, C) Effect of CRISPR/Cas9 editing of E1 and E2 on HES5 and HANR1 expression, as determined by RT-PCR. In (B), HES1 and HANR1 expression was determined in bulk CUTLL1 cells transduced singly or in combination with empty lentiviruses (Cas9GFP, Cas9RFP) or lentiviruses carrying guide RNAs specific for E1 (E1RFP) or E2 (E2GFP). In the left panel in (C), RBPJ motifs are in red and mutated sequences identified by Sanger sequencing of single PCR products are in blue. Data are mean ± SEM of independent determinations done in triplicate. Representative results are shown.
