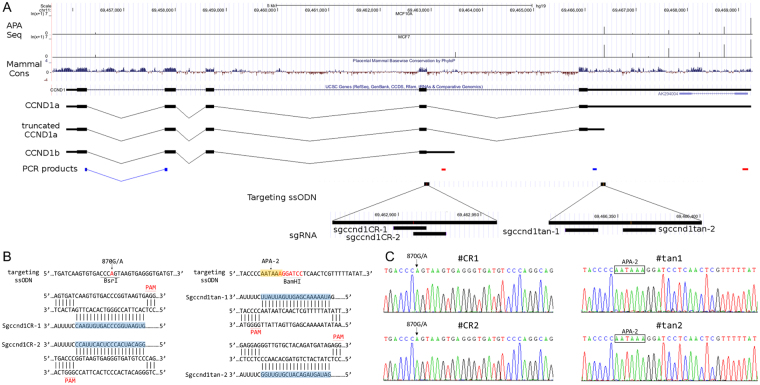
Figure 1.
Alternative polyadenylation of CCND1 and PAS editing with CRISPR/Cas9. (A) Upper panel: APA switching in breast cancer cell lines. MCF10A is a human normal mammary epithelial cell line; MCF7 is a human breast cancer cell line. Lower panel: Schematic representation of the CCND1 locus, APA sites, mRNA isoforms, sgRNA and ssODN. qRT-PCR products used to quantify usage of the APA sites are also shown; the first two correspond to the APA-1 site (CR-APA) and the last two are for the APA-2 site (UTR-APA). Blue represents the common region and red represents the extended region. (B) Sequences of the single-stranded oligonucleotides (ssODN) and sgRNAs used to target the locus. Two sgRNAs were designed for each APA site. Left panel (870 G/A for APA-1): “G” at position 870 is replaced by “A”, which introduces a BsrI site “CCCAGT”; Right panel (APA-2): “AGGATCC” was inserted following “AATAA” at position 304 bp upstream of the stop codon, introducing a canonical PAS “AATAAA” site and a BamHI site. (C) Sequencing validation of the mutated cell lines. #CR1 and #CR2 clones were mutated to use the APA-1 site with sgccnd1CR-1 and sgccnd1CR-2, respectively. #tan1 and #tan2 clones were mutated to use the APA-2 site with sgccnd1tan-1 and sgccnd1tan-2, respectively.