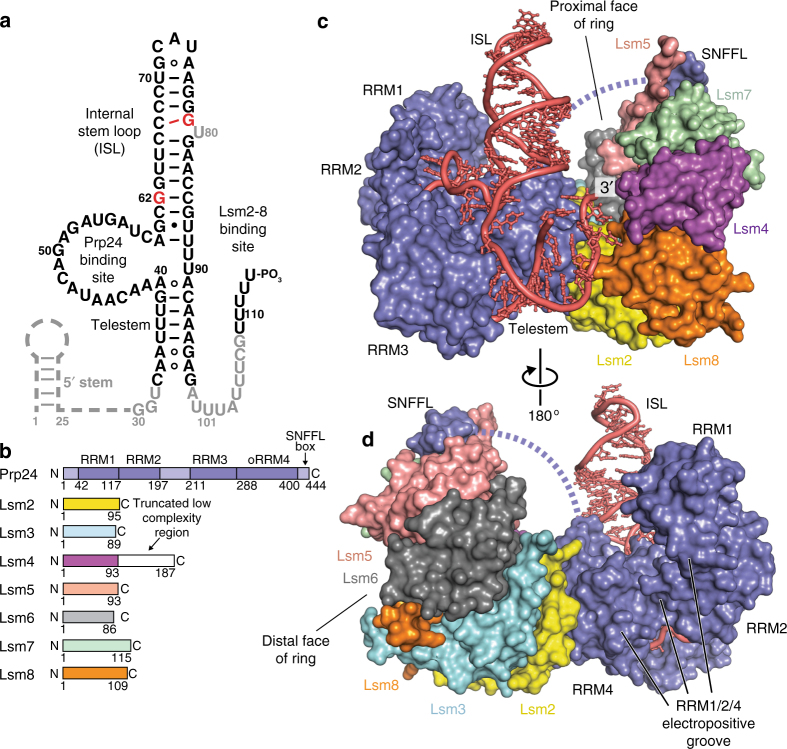
Fig. 1.
Architecture of the U6 snRNP from Saccharomyces cerevisiae. a Sequence and observed secondary structure of the U6 snRNA used for crystallization. Disordered nucleotides are shown in gray text, and the absent 5′ stem region is shown as a dashed line. The 30–113 nucleotide RNA that was used for crystallization has a 3′ phosphate group and A62G/A79G mutations (colored red) to quench structural dynamics in the U6 internal stem loop (ISL), which do not alter the core topology of the U6/Prp24 complex22,23. The 30–112 RNA that yielded a different crystal form contains a 2′ phosphate group and lacks the A79G mutation. b Primary structure of protein components in U6 snRNP samples used for crystallization. The occluded (o) RRM4 of Prp24 has additional N- and C-terminal alpha helices appended to its RRM core76, and is denoted as RRM4 hereafter. In PDB 5VSU, Prp24 residues 1–26 and 399–431 are disordered. In PDB 6ASO, Prp24 residues 1–27 are truncated; 28–29 and 399–444 are disordered. The C-terminus of Lsm4 is predicted to be disordered and was therefore truncated from the crystallization construct (Supplementary Note 1). c Overall architecture of the yeast U6 snRNP, as observed in two distinct crystal structures (Supplementary Table 1). U6 snRNA (red) and Prp24 (blue) contact the “proximal” face of the Lsm2-8 ring. d Alternate view of the U6 snRNP. The ~ 30-residue linker region between RRM4 and the C-terminal SNFFL box motif of Prp24 is disordered and depicted as a dashed line