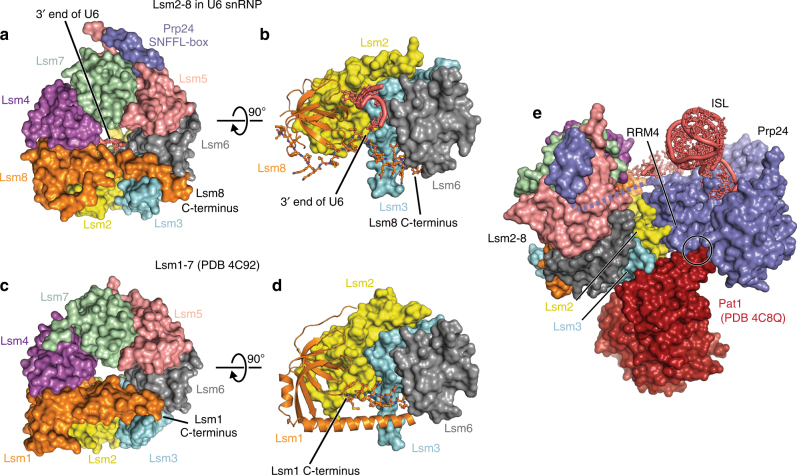
Fig. 2.
Comparison of the Lsm2–8 and Lsm1–7 rings. a The C-terminal region of Lsm8 spans the distal face of the Lsm2–8 ring, contacts the 3′ end of U6 snRNA inside the ring, and forms a C-terminal alpha helix that contacts Lsm3 and Lsm6. b Detailed view of Lsm8 and the 3′ end of U6 snRNA. Lsm4, Lsm5, and Lsm7 are hidden for clarity. Lsm8 is shown in cartoon, except for the C-terminal 34 residues which are shown in ball and sticks. c The C-terminal region of Lsm1 also crosses the distal face of the Lsm1–7 ring via a longer alpha helix that terminates between Lsm3 and Lsm6 before being threaded back into the interior of the ring. d Detailed view of Lsm1 in the Lsm1–7 ring, depicted as in panel c. The C-terminal 12 residues of Lsm1 are shown in ball and sticks. e Superposition of Lsm2–8 in the U6 snRNP with Lsm1–7 bound to RNA decay factor Pat1. The Pat1 binding site on Lsm2 and Lsm3 is solvent exposed in U6 snRNPs. With the exception of a small steric clash between RRM4 and Pat1 (denoted by a circle), the architecture of the U6 snRNP appears to be compatible with binding of Pat1 to Lsm2 and Lsm3