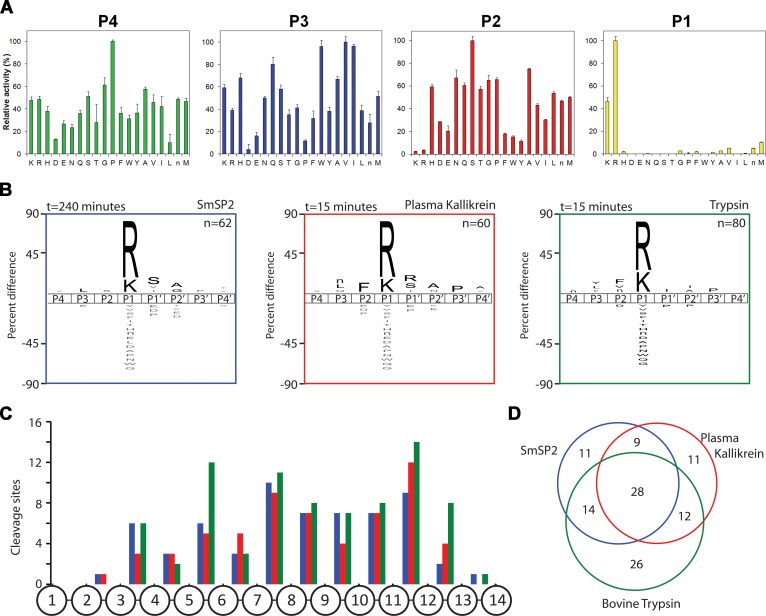
Fig 5. Substrate specificity of rSmSP2.
(A) The P1 to P4 specificity of rSmSP2 was determined by a positional scanning-synthetic combinatorial library. The X-axis indicates 20 amino acids held constant at each position (n is norleucine). The Y-axis represents activity related to the most preferred amino acid (100%). (B) The P4 to P4′ specificity profiles of rSmSP2, human plasma kallikrein, and bovine trypsin were obtained using a multiplex combinatorial library (MSP-MS). The iceLogo substrate profiles were generated from the pattern of cleavage events after incubation with the 14-mer library. Amino acids that are most frequently found at each position are shown above the horizontal line, whereas amino acids that less frequently observed are below. (C) Spatial distribution of cleavage sites within the 14-mer peptide scaffold. (D) The Venn diagram shows the number of unique and shared cleavage sites.