Fig. 5. Posttranslational modification sites on HupB.
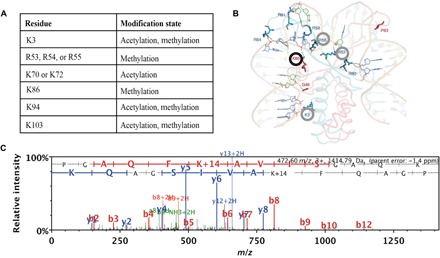
(A) Modified amino acids and posttranslational modification sites on HupB identified by whole-cell proteomics. (B) Predicted structure of M. tuberculosis HupB reprinted with permission from Bhowmick et al. (30). Modified amino acid residues identified in this study are circled. The K86 residue, the spectrum of which is depicted in (C), is circled in black. (C) MS/MS of tryptic peptide identified after a Mascot database search, where methylation on Lys86 can be inferred from the peptide mass/charge ratio (m/z) and the ion fragmentation pattern.
