Fig. 2. HGT accounts for the presence of ωx desaturase in some animal genomes.
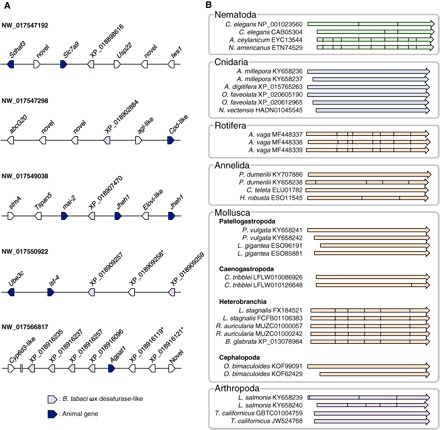
(A) Multiple copies of ωx desaturase–like sequences scattered among several loci of the B. tabaci genome. The ωx desaturase–like sequences are found in five different scaffolds and are flanked by animal genes. * indicates sequences that did not pass the filter (probability between 0.50 and 0.70). (B) Intron-exon structures in the ωx desaturase genes from selected metazoan species. The arrows indicate aligned coding sequences (CDS) extracted from the genomic assembly of each species. The positions of introns are indicated as solid lines within the arrows. The dotted line indicates shared intron-exon boundaries. All cnidarian genes and several genes from Annelida and Mollusca have an “intronless” organization. The CDS of the three ωx desaturase genes from A. vaga (Rotifera) share the same overall structure consisting of eight exons. In addition, all genes from three Heterobranchia species have the same structure with six exons in the CDS. Variable gene structures were found in Nematoda, Annelida, and Arthropoda. The arrow colors are consistent with those used to represent clades in Fig. 1.
