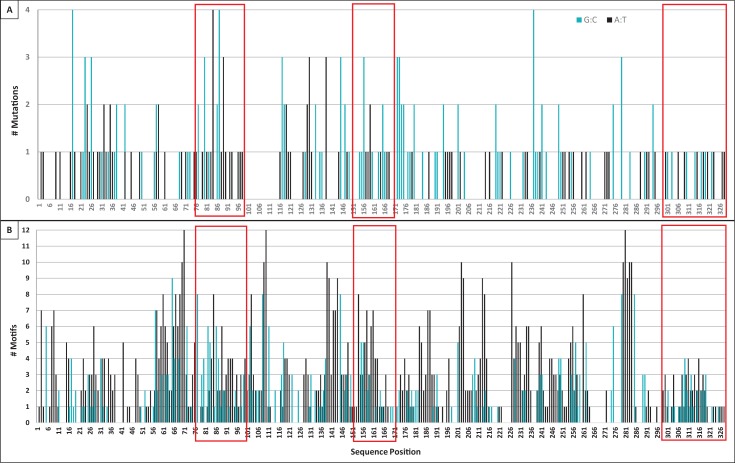
Figure 7. Mutation and motif locations within individual domains of TcR αV sequences.
(A) Number of mutations (both single and tandem, 283 total) to A:T nucleotides (black bars, 122 mutations) or G:C nucleotides (blue bars, 161 mutations) observed at each position along the sequence length of TcR αV sequences. We counted mutations to Geneious-derived consensus sequences within framework and complementarity-determining region (CDR) domains (41 sequences within 11 αV groups). (B) Number of WA/TW motifs (black bars, indicating possible polymerase η action) or DGYW/WRCH motifs (blue bars, indicating hotspots for AID activity, and thus possible mutation) at each position along the sequence length. Position indicates the forward (3’ to 5’) location of the mutable base of each motif within a Geneious-derived consensus sequence for each TcR αV group (eleven groups). Red boxes indicate the location of CDR1 (positions 76–99), CDR2 (positions 151–171), and CDR3 (positions 300–328) within each panel. [WA/TW: A:T is the mutable position; DGYW/WRCH: G:C is the mutable position; W = A/T, D = A/G/T, Y = C/T, R = A/G, and H = T/C/A].