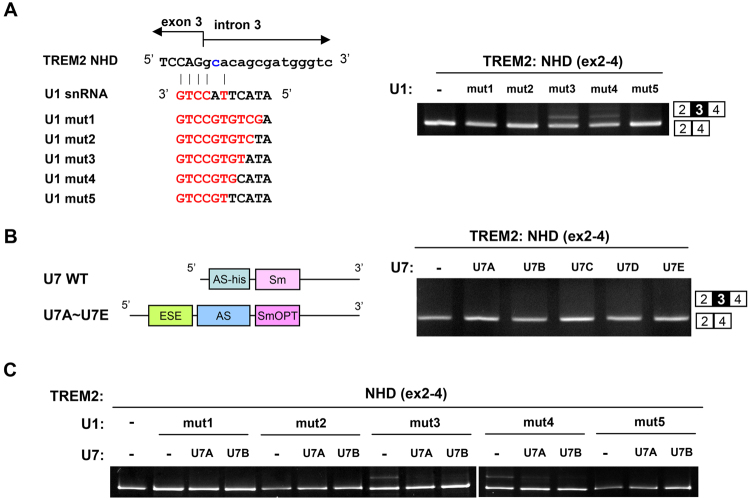
Figure 2.
Correction of exon 3 skipping by modified U1 and U7 snRNAs. (A) The left panel shows the sequence of the 5′ region of the unmodified (WT) and modified U1 snRNAs used in this study. The mutated nucleotide in the NHD minigene is shown in blue. Nucleotides of the U1 snRNA constructs complementary to the 5′ splice site region of TREM2 intron 3 are indicated in red. The right panel shows the results of a splicing assay using modified U1 snRNA. The NHD minigene was transfected with either an empty vector or modified constructs. Splicing patterns were detected by RT-PCR and agarose gel electrophoresis. U1mut3 increased exon 3 inclusion. (B) The left panel shows a schematic diagram of the modified U7 snRNA used in this study. U7A–U7E were designed to hybridize to different regions in exon 3 of TREM2. Refer to the text for the details regarding SmOPT and ESE (exonic splicing enhancer). The right panel shows the results of a splicing assay using the modified U7 snRNA. The NHD minigene was transfected with either an empty vector or modified constructs. Splicing patterns were detected as in (A). U7A and U7B did not increase exon 3 inclusion when used separately or simultaneously. (C) Co-expression of U1 and U7 constructs to modulate exon 3 splicing. Cells were transfected with the NHD minigene together with the U1 and U7 constructs as indicated. Splicing patterns were detected as in (A). Original gel images are shown in Supplementary Fig. S8.