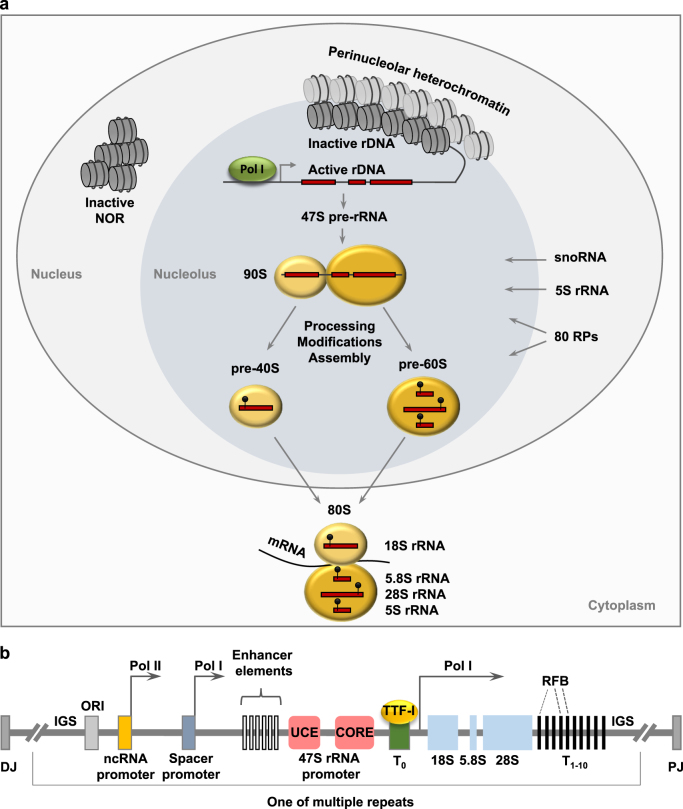
Fig. 1.
Nucleolus and rDNA genes. a Intranucleolar heterochromatic inactive rDNA repeats are tightly connected with perinucleolar heterochromatin, whereas euchromatic active rDNA genes are transcribed into the 47 S pre-rRNA by Pol I. The 47 S pre-rRNA is co-transcriptionally assembled into the 90 S processome with RPs and 5 S rRNA and modified by ~200 snoRNPs. Following cleavage of the pre-rRNA, the 90 S processome separates into a pre-60S subunit and a pre-40S subunit that largely follow independent maturation pathways in the nucleolus, the nucleoplasm and the cytoplasm. Modifications of rRNA are indicated with lollipops. b Schematic diagram of the rDNA array. Shown are coding regions for 18 S, 5.8 S, and 28 S rRNA as well as IGS with the rDNA core promoter and upstream control element (UCE), enhancers, spacer promoter, and Pol I promoter, ORIs as well as T0 that binds TTF-I and T1, T4, and T5 that act as RFBs. DJ and PJ sequences that flank the rDNA array are indicated