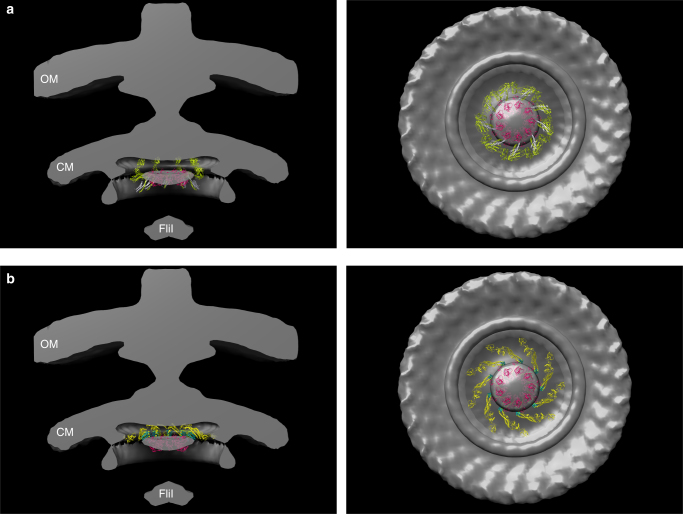
Fig. 6.
Fitting of the FlhAC ternary complexes into the cryoEM density map of the flagellum base body44. The nonameric oligomeric structure of FlhAC was generated using the structure of the homologous MxiA protein28. The ternary structures obtained in this work were then used to model the complete oligomeric nonameric ring of FlhAC bound to nine molecules of a FliT−FliD and b FliS−FliC. Non-cooperative binding of the chaperone–substrate molecules on FlhAC is assumed. The full-length FliD (5H5V) and FliC (PDB ID 3A5X) structures were used. FliI is the hexameric flagellar ATPase. CM cytoplasmic membrane; OM outer membrane. Side views of half-cut sections and bottom views are shown. The colors of the FlhAC-chaperone–substrate complexes are as in Figs. 2 and 3