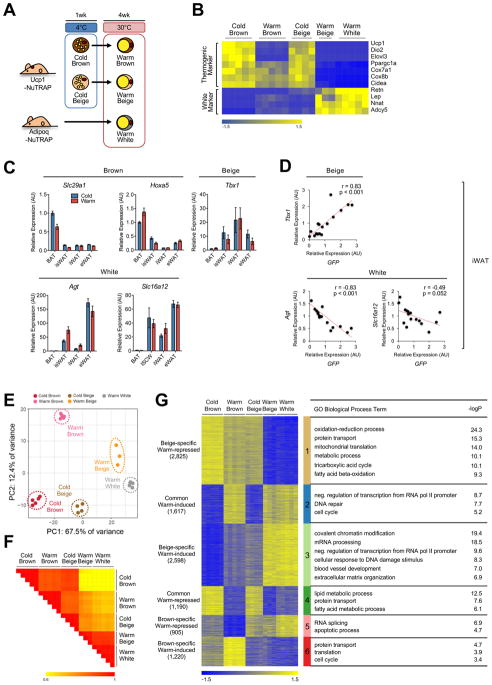
Figure 2. Beige and brown adipocytes exhibit distinct transcriptomic changes upon warming.
(a) Experimental scheme showing how different types of adipocytes are collected. Brown and beige adipocytes (cold/warm) were collected from BAT and iWAT of Ucp1-NuTRAP mice, respectively, exposed to cold and after 4 weeks of warming at 30°C. White adipocytes were collected from iWAT of Adipo-NuTRAP mice housed at thermoneutrality from birth.
(b) Heatmap of marker gene expression for different adipocyte types from cold and warm conditions as described in (a). Columns represent biological replicates. Expression values (Counts Per Million; CPM) of each mRNA are represented by z-scores.
(c) Gene expression analysis by qRT-PCR of proposed brown, beige and white adipocyte markers in different fat depots (whole tissues) in response to cold (4°C, 1 week) and subsequent warming (30°C, 4 weeks). Bars indicate mean ± SEM (n=6 animals/group). BAT, interscapular brown adipose tissue; isWAT, interscapular white adipose tissue; iWAT, inguinal white adipose tissue; eWAT, epididymal white adipose tissue.
(d) Correlation analysis between expression of beige/white adipocyte markers and GFP within iWAT. Each dot indicates an individual sample.
(e) PCA of transcriptome of adipocyte types in cold and warm conditions. Each dot indicates an individual biological replicate (n=3–5 animals/replicate).
(f) Similarity matrix showing pairwise Pearson correlations for RNA-seq profiles. Pearson correlation coefficient r is represented in color as indicated.
(g) Heatmap of K-mean (K=6) clustering of transcriptomic profiles for differential genes. The number of genes in each cluster is in parentheses; each cluster is indicated by a different vertical color bar. Pathways (GO biological process) enriched in each cluster and their corresponding −log10 p-values are shown in the table at right.