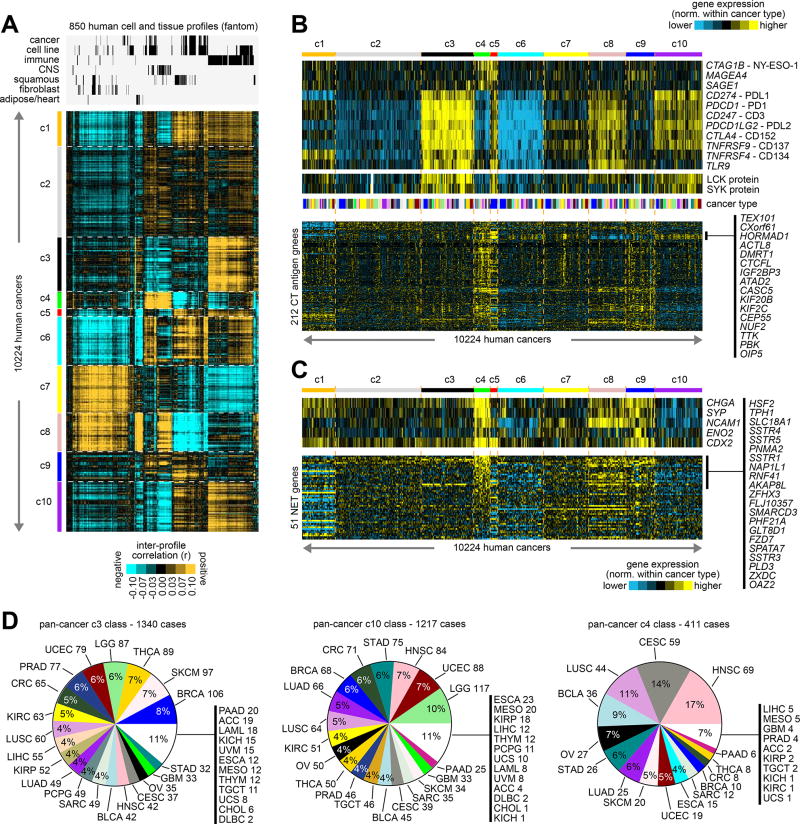
Figure 3. Normal tissue and cell type associations with the pan-cancer molecular classes.
(A) Inter-profile correlations were computed between TCGA expression profiles (with values normalized within each cancer type) and profiles from the Fantom consortium expression dataset of various cell types or tissues from human specimens (n=850 profiles)(10). Membership of the Fantom profiles in general categories of “cancer”, “cell line”, “immune” (immune cell types or blood or related tissues), “CNS” (related to central nervous system including brain), “squamous” (including bronchial, trachea, oral regions, throat and esophagus regions, nasal regions, urothelial, cervix, sebocyte, keratin/skin/epidermis), “fibroblast”, or “adipocyte/heart” is indicated. Cancer type color coding in part D. (B) Heat maps of differential expression (values normalized within each cancer type), for genes encoding immunotherapeutic targets (top), for LCK and SYK proteins (middle, representing markers for T-cells and B-cells, respectively), and for genes encoding cancer-testis (CT) antigens (from the CT Gene Database, http://cancerimmunity.org/resources/ct-gene-database/). (C) Heat maps of differential expression (values normalized within each cancer type), for genes encoding canonical markers of neuroendocrine tumors (top), and for a set of 51 genes in a panel of neuroendocrine tumor (NET) markers(34), as uncovered previously using gene expression profiling (bottom). (D) For c3 (immune-associated), c10 (immune-associated), and c4 (CNS- and neuroendocrine-associated) pan-cancer classes, distributions by cancer type. See also Supplementary Figure 13 and Supplementary Data 5.