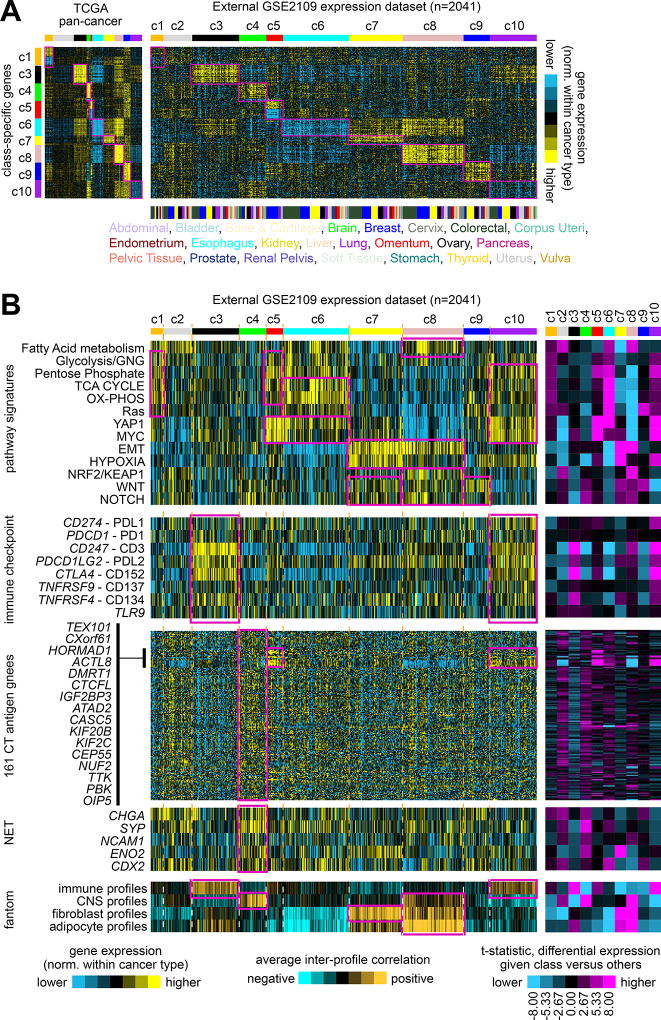
Figure 5. Observation of patterns associated with TCGA pan-cancer molecular classes in an external multi-cancer expression profiling dataset.
(A) Gene expression profiles of 2041 cancer cases of various pathologically defined cancer types, represented in the Expression Project for Oncology (expO) (GSE2109) dataset (profiles being normalized within their respective cancer type), were classified according to TCGA pan-cancer molecular class. Expression patterns for the top set of 854 mRNAs distinguishing between the ten TCGA molecular classes (from Figure 1A) are shown for both TCGA and GSE2109 datasets. Genes in the GSE2109 sample profiles sharing similarity with TCGA class-specific signature pattern are highlighted. (B) In the same manner as carried out for TCGA datasets, expO expression profiles were scored for pathway-associated gene signatures (from Figure 2A), surveyed for immune checkpoint markers and for CT antigen genes (from Figure 3B, using the same gene ordering), surveyed for canonical neuroendocrine tumor (NET) markers (from Figure 3C), and scored for similarity to normal cell type categories represented in the fantom dataset (from Figure 3A). Pan-cancer class associations of particular interest (which tend to follow the patterns first observed in TCGA cohort) are highlighted. The purple-cyan heat maps off to the right denote t-statistics for comparing the given class versus the rest of the tumors; dark purple or cyan corresponds approximately to p<0.01. Parts A and B have the same ordering of expO expression profiles. See also Supplementary Figures 14 to 16 and Supplementary Data 6.