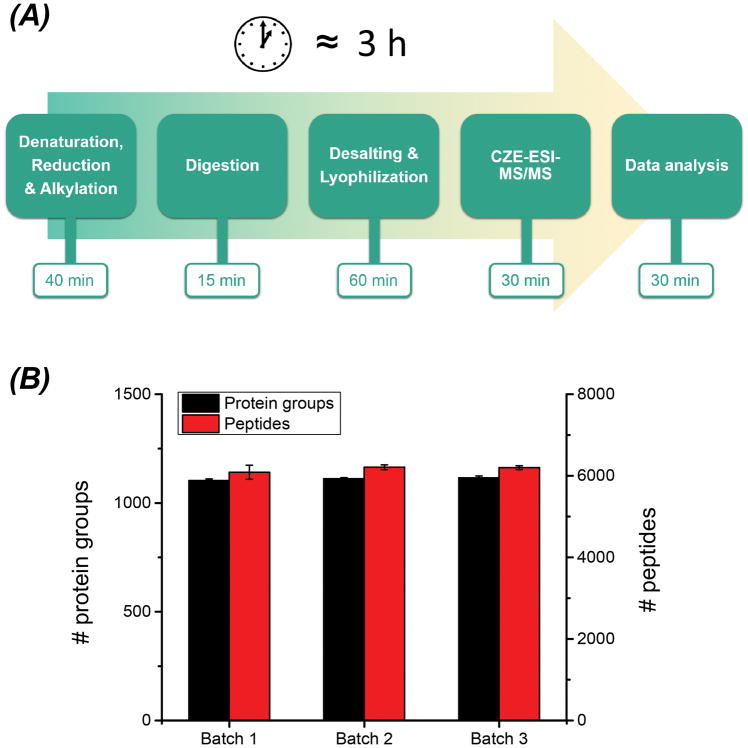
Figure 5.
The high-throughput bottom-up proteomics workflow using IM-N for rapid protein digestion and CZE-MS/MS for fast sample analysis (A). The number of protein and peptide IDs from the mouse brain proteome using the workflow (B). Three samples were prepared and analyzed by the workflow as three batches. Each sample was analyzed by the CZE-MS/MS in triplicate. The error bars represent the standard deviations of the number of protein and peptide IDs from the triplicate CZE-MS/MS analyses.