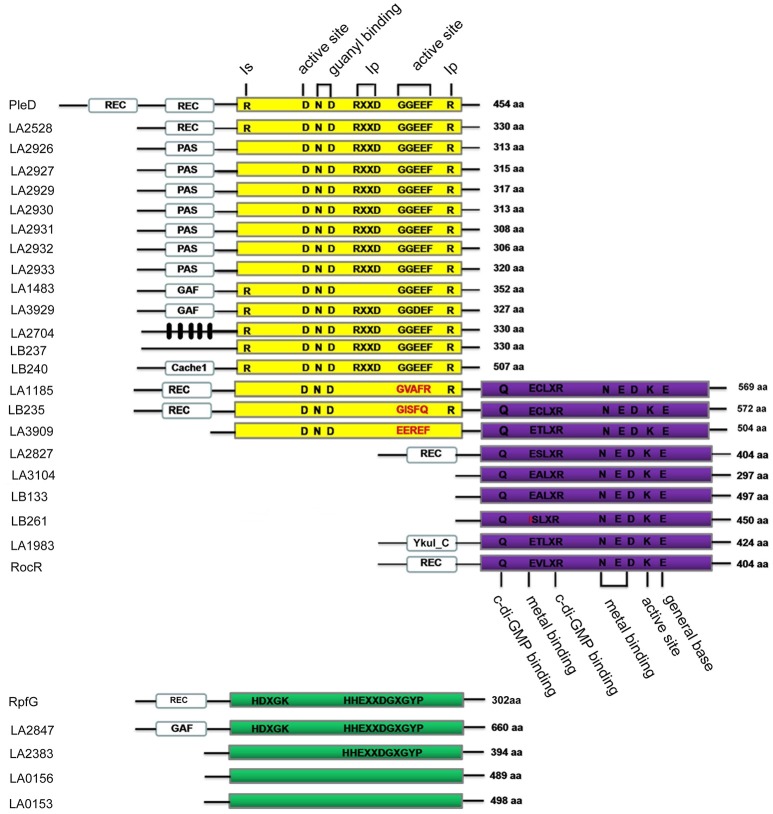
Figure 1.
Schematic representation of the domain architecture of c-di-GMP metabolic enzymes of L. interrogans Lai strain 56601. Protein names are marked on the left and the number of amino acids is shown on the right. Yellow, purple, and green boxes represent GGDEF domains, EAL domains and HD-GYP domains, respectively. The GGDEF domain protein PleD from C. Crescentus, the EAL domain protein RocR from P. Aeruginosa and the HD-GYP domain protein RpfG from X. campestris are used as references to highlight amino acids important for enzymatic activity (Ryan et al., 2006; Wassmann et al., 2007; Rao et al., 2008). The putative conserved functional amino acid residues are shown in black (conserved) or red (mutation). Black boxes are predicted transmembrane regions. Ip is the primary inhibitory binding site for c-di-GMP, and Is is the secondary inhibitory binding site for c-di-GMP. REC, PAS, Cache1, GAF, and YkuI_C sensory domains are indicated by white boxes.