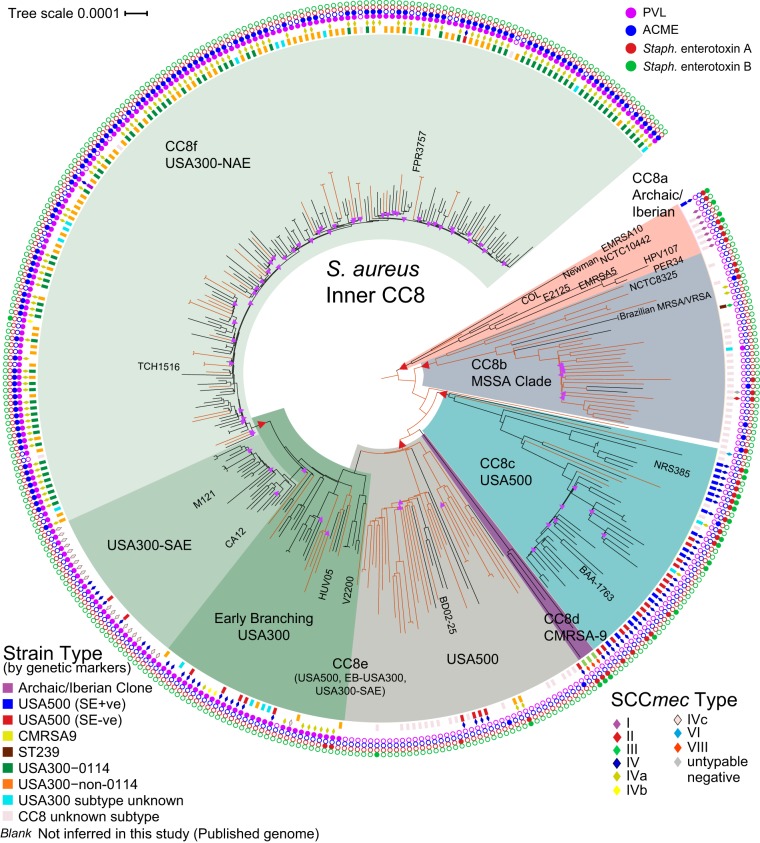
FIG 2 .
WGS-based maximum likelihood phylogeny (using the best-fit model TVMe+ASC) of 348 genomes of S. aureus (229 MRSA and 119 MSSA) belonging to the Inner CC8 clade (excluding ST239 and ST630 genomes), illustrating the relationship structure of clinically important CC8 groups and showing that genetic marker inference (GMI) strain typing is not always indicative of genetic relationship. MSSA genomes, on branches colored orange (Hex DF5000, RGB [223,80,0]), are interspersed among MRSA genomes. This analysis includes 1,000 bootstraps of 13,988 SNPs. Nodes with bootstrap values of <90% are marked with purple triangles. The core genome size is 2.26 Mbp (78.8% of reference genome FPR3757). Branches of the phylogeny on which SNPs were selected for assay development are marked with a red triangle.