Table 2.
Drug targets (genes/proteins), their products and functional activities and mode of action of anti-TB drugs.
| Drug targets | Size (bp) | Protein length | Molecular mass (Da) | Product | Functional activity | Mechanisms of anti-TB drugs | Structure & Pubchem CID | References |
|---|---|---|---|---|---|---|---|---|
| katG (Rv1908c) | 2,223 | 740 | 80,572.8 | Catalase-peroxidase | Intracellular survival of mycobacteria |
INH: Inhibition of mycolic acid synthesis Activity: Bactericidal |
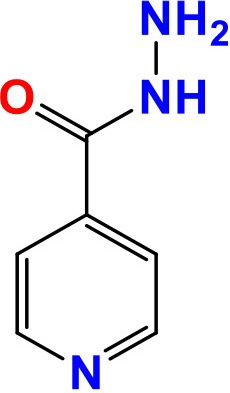 Pubchem CID: 3767 |
Ramaswamy et al., 2003; Vilchèze et al., 2006, 2007; Forrellad et al., 2013; Machado et al., 2013; Bantubani et al., 2014; Seifert et al., 2015; Zhang and Yew, 2015; Aye et al., 2016; Datta et al., 2016 |
| InhA (Rv1484) | 810 | 269 | 28,527.8 | NADH-dependent enoyl ACP reductase | Mycolic acid biosynthesis | |||
| AhpC (Rv2428) | 588 | 195 | 21,566.3 | Alkyl hydroperoxide reductase | Protection against oxidative stress | |||
| FabG1 (Rv1483) | 744 | 247 | 25,665.2 | 3-Oxoacyl (acyl-carrier protein) reductases | Fatty acid biosynthesis pathway | |||
| iniA (Rv0342) | 1,923 | 640 | 70,082.9 | INH inductible gene protein IniA | Efflux pump associated | |||
| FadE24 (Rv3139) | 1,407 | 468 | 49,646.4 | acyl-CoA dehydrogenase FadE24 | Involved in lipid degradation and fatty acid β –oxidation | |||
| KasA (Rv2245) | 1,251 | 416 | 43,284.1 | β -Ketoacyl acyl carrier protein synthase | Involved in fatty acid biosynthesis | |||
| ndh (Rv1854c) | 1,392 | 463 | 49,619 | NADH dehydrogenase Ndh | Electrons transference from NADH to the respiratory chain | |||
| rpoB (Rv0667) | 3,519 | 1172 | 129,218 | β-subunit of RNA polymerase | Catalyzes the transcription of DNA into RNA |
RIF: Inhibition of RNA synthesis Activity: Bactericidal |
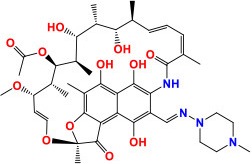 Pubchem CID: 5458213 |
Mboowa et al., 2014; Ocheretina et al., 2014; Thirumurugan et al., 2015; Aye et al., 2016 |
| rpoA (Rv3457c) | 1,044 | 347 | 37,706.5 | α-subunit of RNA polymerase | Catalyzes the transcription of DNA into RNA | |||
| rpoC (Rv0668) | 3,951 | 1316 | 146,737 | β′-subunit of RNA polymerase | Catalyzes the transcription of DNA into RNA | |||
| pncA (Rv2043c) | 561 | 186 | 19,604.6 | Pyrazinamidase/nicotinamidase (PZase) | Converts amides into acid (PZA into POA) |
PZA: Inhibition of trans-translation and pantothenate and CoA synthesis; depletes of membrane energy Activity: Bactericidal / bacteriostatic |
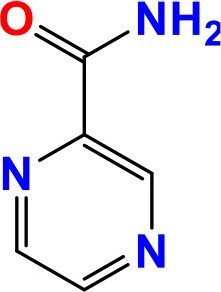 Pubchem CID: 1046 |
Juréen et al., 2008; Shi et al., 2011b, 2014; Njire et al., 2016, 2017; Zhang et al., 2017 |
| RpsA (Rv1630) | 1,446 | 481 | 53,201.8 | 30S ribosomal protein S1 | Translate mRNA with a shine-dalgarno (SD) purine-rich sequence | |||
| panD (Rv3601c) | 420 | 139 | 14,885 | Aspartate alpha-decarboxylase | Pantothenate biosynthesis | |||
| clpC1 (Rv3596c) | 2,547 | 848 | 93,552.4 | ATP-dependent protease ATP-binding subunit ClpC1 | Protein degradation, Hydrolyses proteins in presence of ATP | |||
| gpsI (Rv2783c) | 2,259 | 752 | 79,734.7 | Bifunctional protein polyribonucleotide nucleotidyltransferase GpsI (pnpase) | Involved in mRNA degradation | |||
| rpsL (Rv0682) | 375 | 124 | 13,849.2 | 30S ribosomal protein S12 RpsL | Translation initiation step |
STR: Inhibition of protein synthesis Activity: Bactericidal |
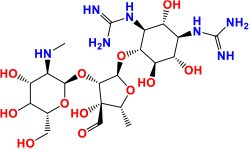 Pubchem CID: 19649 |
Finken et al., 1993; Sharma et al., 2007; Chakraborty et al., 2013; Du et al., 2013; Perdigão et al., 2014; Smittipat et al., 2016 |
| rrs (MTB000019) | 1,537 | — | — | 16S ribosomal RNA | Stable RNAs | |||
| gidB (Rv3919c) | 675 | 224 | 24,031.9 | 7-methylguanosine methyltransferase | Probable glucose-inhibited division protein B Gid | |||
| EmbB (Rv3795) | 3,297 | 1098 | 118,021 | Arabinosyl transferase EmbB | Biosynthesis of the mycobacterial cell wa |
EMB: Inhibition of arabinogalactan synthesis Activity: Bacteriostatic |
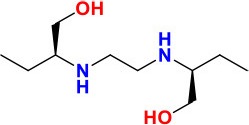 Pubchem CID: 14052 |
Mikusová et al., 1995; Wang et al., 2010; Safi et al., 2013; Yoon et al., 2013; Moure et al., 2014; Zhang et al., 2014; Brossier et al., 2015; Xu et al., 2015 |
| EmbA (Rv3794) | 3,285 | 1094 | 115,692 | Arabinosyl transferase EmbA | Biosynthesis of the mycobacterial cell wall | |||
| EmbC (Rv3793) | 3,285 | 1094 | 117,490 | Arabinosyl transferase EmbC | Biosynthesis of the mycobacterial cell wall | |||
| embR (Rv1267c) | 1,167 | 388 | 41,933.5 | Transcriptional regulatory protein EmbR | Regulator of embCAB operon transcription | |||
| rmlD (Rv3266c) | 915 | 304 | 32,044.8 | dTDP-4-dehydrorhamnose reductase | Involved in dTDP-L-rhamnose biosynthesis | |||
| gyrA (Rv0006) | 2,517 | 838 | 92,274.3 | DNA gyrase subunit A | Negatively supercoils closed circular double-stranded DNA |
Fluoroquinolone: Inhibition of DNA synthesis Activity: Bactericidal |
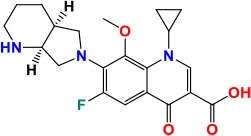 Moxifloxacin Pubchem CID: 152946 |
Aubry et al., 2004; Von Groll et al., 2009; Cui et al., 2011; Nosova et al., 2013; Li et al., 2014 |
| gyrB (Rv0005) | 2,142 | 675 | 74,058.7 | DNA gyrase subunit B | Negatively supercoils closed circular double-stranded DNA | |||
| rrs (MTB000019) | 1,537 | — | — | 16S ribosomal RNA | Stable RNAs |
KAN/AMK/CAP/VIM: Inhibition of protein synthesis Activity: Bactericidal |
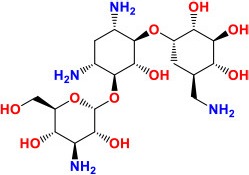 Kanamycin Pubchem CID: 6032 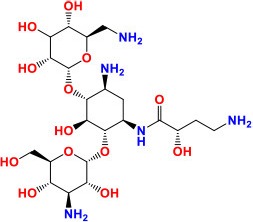 Amikacin Pubchem CID: 37768 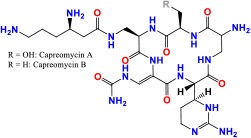 Capreomycin Pubchem CID: 3000502 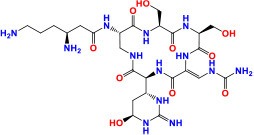 Viomycin Pubchem CID: 3037981 |
Stanley et al., 2010; Ajbani et al., 2011; Yuan et al., 2012; Du et al., 2013; Reeves et al., 2013; Sowajassatakul et al., 2014; Freihofer et al., 2016 |
| eis (Rv2416c) | 1,209 | 402 | 43,771.8 | Aminoglycoside N-acetyltransferase | Acetylation, intracellular survival | |||
| whiB7 (Rv3197A) | 279 | 92 | 10,106.7 | Transcriptional regulatory protein WhiB7 | Transcriptional mechanism | |||
| TlyA (Rv1694) | 807 | 268 | 28,042.1 | 2′-O-methyltransferase | Methylates 16S and 23S rRNA | |||
| InhA (Rv1484) | 810 | 269 | 28,527.8 | NADH-dependent enoyl ACP reductase | Mycolic acid biosynthesi |
ETO: Disrupts cell wall biosynthesis Activity: Bacteriostatic |
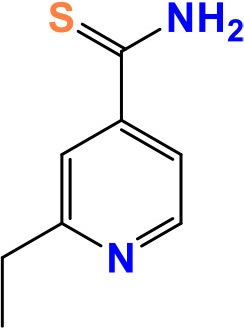 Pubchem CID: 2761171 |
Zhang et al., 1992; Vilchèze et al., 2006, 2008; Carette et al., 2011; Grant et al., 2016; Thee et al., 2016; Mori et al., 2017 |
| ethA (Rv3854c) | 1,470 | 489 | 55,326.1 | Monooxygenase EthA | Activates the pro-drug ethionamide (ETH) | |||
| ethR (Rv3855) | 651 | 216 | 23,724.7 | Transcriptional repressor protein EthR | Regulates negatively the production of ETHA | |||
| KasA (Rv2245) | 1,251 | 416 | 43,284.1 | β -Ketoacyl acyl carrier protein synthase | Involved in fatty acid biosynthesis | |||
| thyA (Rv2764c) | 792 | 263 | 29,820.8 | Thymidylate synthase ThyA | Deoxyribonucleotide biosynthesis |
PAS: Inhibits of folic acid and thymine nucleotide metabolism Activity: Bacteriostatic |
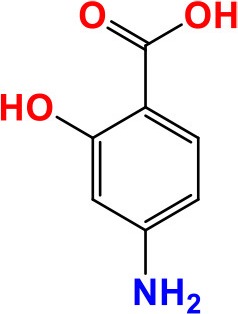 PubchemCID: 4649 |
Rengarajan et al., 2004; Mathys et al., 2009; Zhao et al., 2014; Meumann et al., 2015 |
| folC (Rv2447c) | 1,464 | 487 | 50,779.3 | Folylpolyglutamate synthase protein FolC | Conversion of folates to polyglutamate derivatives | |||
| dfrA (Rv2763c) | 480 | 159 | 17,640 | Dihydrofolate reductase DfrA | For de novo glycine and purine synthesis | |||
| RibD (Rv2671) | 777 | 258 | 27,693.5 | Bifunctional enzyme riboflavin biosynthesis protein RibD | Involved in riboflavin biosynthesis | |||
| alr (Rv3423c) | 1,227 | 408 | 43,355.6 | Alanine racemase Alr | Provides D-alanine required for cell wall biosynthesis |
DCS: Inhibits the synthesis of peptidoglycan in the cell wall Activity: Bacteriostatic |
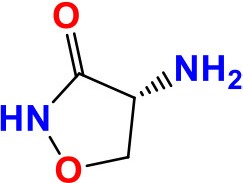 PubchemCID: 6234 |
Cáceres et al., 1997; Andini and Nash, 2006; Chen et al., 2012; Zhang and Yew, 2015; Desjardins et al., 2016 |
| ddl (Rv2981c) | 1,122 | 373 | 39,678.2 | D-alanine: D-alanine ligase DdlA | Involved in cell wall formation | |||
| Ald (Rv2780) | 1,116 | 371 | 38,713.2 | L-alanine dehydrogenase | Cell wall synthesis | |||
| cycA (Rv1704c) | 1,671 | 556 | 60,047.2 | D-serine / alanine / glycine transporter protein CycA | transport across the cytoplasmic membrane | |||
| Rv0678 | 498 | 165 | 18,346.7 | Conserved protein | Transcription repressor for efflux pump MmpL5 |
CLO: Produces reactive oxygen, inhibits energy production, memberane distruption Activity: Bacteriostatic |
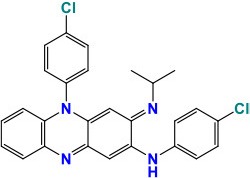 Pubchem CID: 2794 |
Huitric et al., 2010; Yano et al., 2011; Hartkoorn et al., 2014; Almeida et al., 2016 |
| Rv1979c | 1,446 | 481 | 51,084.1 | Conserved permease | Probably involved in transportation of amino acid across the membrane. | |||
| pepQ (Rv2535c) | 1,119 | 372 | 38,758.9 | Cytoplasmic peptidase PepQ | Possibly hydrolyses peptides | |||
| rplC (Rv0701) | 654 | 217 | 23,090.5 | Encodes the 50S ribosomal L3 protein | Formation of peptidyltransferase center of the ribosome |
LIN/SZD: Both act by fixation of an early step in protein synthesis Activity: Bactericidal |
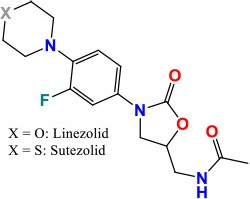 Pubchem CID: 441401 |
Williams et al., 2009; Beckert et al., 2012; Makafe et al., 2016; Zhang et al., 2016 |
| rrl (MTB000020) | 3,138 | —– | —– | Ribosomal RNA 23S | Stable RNAs | |||
| Ddn (Rv3547) | 456 | 151 | 17,371 | Deazaflavin-dependent nitroreductase | Converts bicyclic nitroimidazole drug candidate pa-824 to three metabolites, |
DMD: Impeding the synthesis of mycolic acid Activity: Bactericidal |
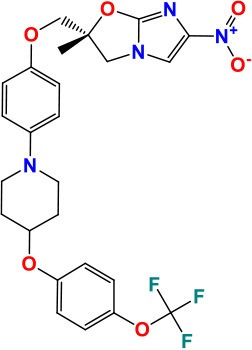 Pubchem CID: 6480466 |
Matsumoto et al., 2006; Palomino and Martin, 2014; Shimokawa et al., 2014; Bloemberg et al., 2015; Haver et al., 2015 |
| Fgd1 (Rv0407) | 1,011 | 336 | 490.783 | F420-dependent glucose-6-phosphate dehydrogenase | Catalyzes oxidation of glucose-6-phosphate to 6-phosphogluconolactone using coenzyme F420 as the electron acceptor | |||
| fbiA (Rv3261) | 996 | 331 | 3,640.54 | F420 biosynthesis protein FbiA | Required for coenzyme F420 production: involved in the conversion of FO into F420 | |||
| fbiB (Rv3262) | 1,347 | 448 | 3,641.53 | F420 biosynthesis protein FbiB | Required for coenzyme F420 production: involved in the conversion of FO into F420 | |||
| fbiC (Rv1173) | 2,571 | 856 | 1,302.93 | F420 biosynthesis protein FbiC | Participates in a portion of the F420 biosynthetic pathway between pyrimidinedione and FO | |||
| mmpL3 (Rv0206c) | 2,835 | 944 | 100,872 | Conserved transmembrane transport protein MmpL3 | Unknown; Supposed to be involved in fatty acid transport |
SQ-109: Acts by interfering with the assembly of mycolic acids into the bacterial cell wall core Activity: Bactericidal |
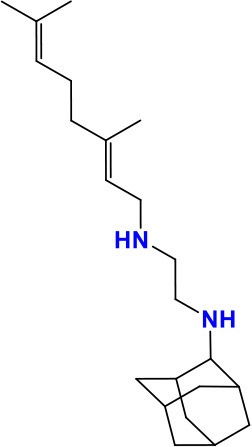 Pubchem CID: 5274428 |
Boshoff et al., 2004; Jia et al., 2005; Grzegorzewicz et al., 2012; |
| DprE1 (Rv3790) | 1,386 | 461 | 50,163.2 | Decaprenylphosphoryl-beta-D-ribose 2′-oxidase | Together with DPRE2 arabinan synthesis |
BTZ: Drug is activated by reduction of an essential nitro group to a nitroso derivative, which can react with a cysteine residue in DprE1 Activity: Bactericidal |
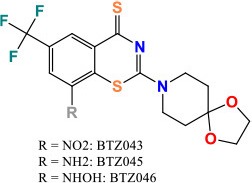 ChemSpider ID: 25941868 |
Manina et al., 2010; Trefzer et al., 2010; Kolly et al., 2014; Makarov et al., 2014 |
| DprE2 (Rv3791) | 765 | 254 | 27,468.8 | Decaprenylphosphoryl-D-2-keto erythro pentose reductase | Together with DPRE1 arabinan synthesis |
