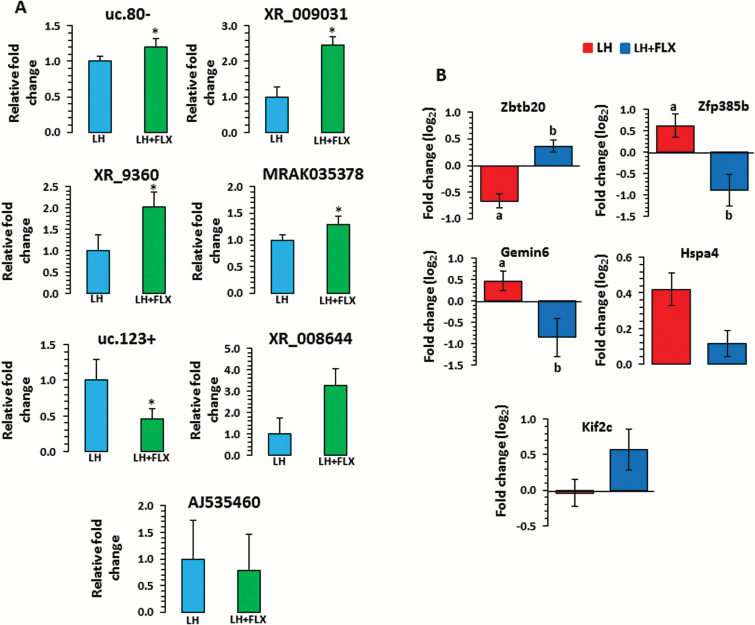
Figure 4.
qPCR-based expression-associated changes in long noncoding RNAs (lncRNAs) and target genes in learned helplessness (LH) rats treated with fluoxetine (LH+FLX). (A) Transcript levels of uc.80-, XR_009031, XR_9360, MRAK035378, uc.123+, XR_008644, and AJ535460 were analyzed in LH rats tested under fluoxetine treatment by qPCR using primers mentioned in the Methods section. Gapdh normalized expression level for each transcript is presented as relative fold change. All data demonstrate the mean±SEM (for uc.80, n=5 in LH, n=5 in LH+FLX; for XR_009031, n=5 in LH, n=7 in LH+FLX; for XR_9360, n=4 in LH, n=6 in LH+FLX; for MRAK035378, n=6 in LH, n=7 in LH+FLX; for uc.123+, n=4 in LH, n=5 LH+FLX; for XR_008644, n=7 in LH, n=7 in LH+FLX and for AJ535460, n=6 in LH, n=5 in LH+FLX). The level of significance was determined using independent-sample t test. *Significant difference between 2 compared groups (for uc.80-, P=.03; XR_009031, P=.003; XR_9360, P=.04; MRAK035378, P=.04; uc.123+, P=.003; XR_008644, P=.07 and AJ535460, P=.35). (B) Transcript levels of target genes (log2 fold change) including Zbtb20, Zfp385b, Gemin6, Hspa4, and Kif2c were analyzed in fluoxetine-treated LH (LH+FLX) rats and compared with untreated LH rats. All data are the mean±SEM (for Zbtb20, n=6 in LH and n=7 in LH+Flx; Zfp385b, n=5 in LH and n=4 in LH+Flx; for Gemin6, n=5 in LH and n=5 in LH+Flx; Hspa4, n=7/group and Kif2c, n=5 in LH and n=5 in LH+Flx). The level of significance was determined using independent-sample t test. a denotes significant difference between untreated LH and TC groups (for Zbtb20: P=.0009; Zfp385b: P=.02; and Gemin6: P=.03), whereas b denotes significant difference between LH+FLX and untreated LH groups (for Zbtb20, P=.02; Zfp385b, P=.04; and Gemin6, P=.05). On the contrary, changes associated with Hsp4 and Kif2c were not significant in both LH vs TC and LH+FLX vs LH groups.