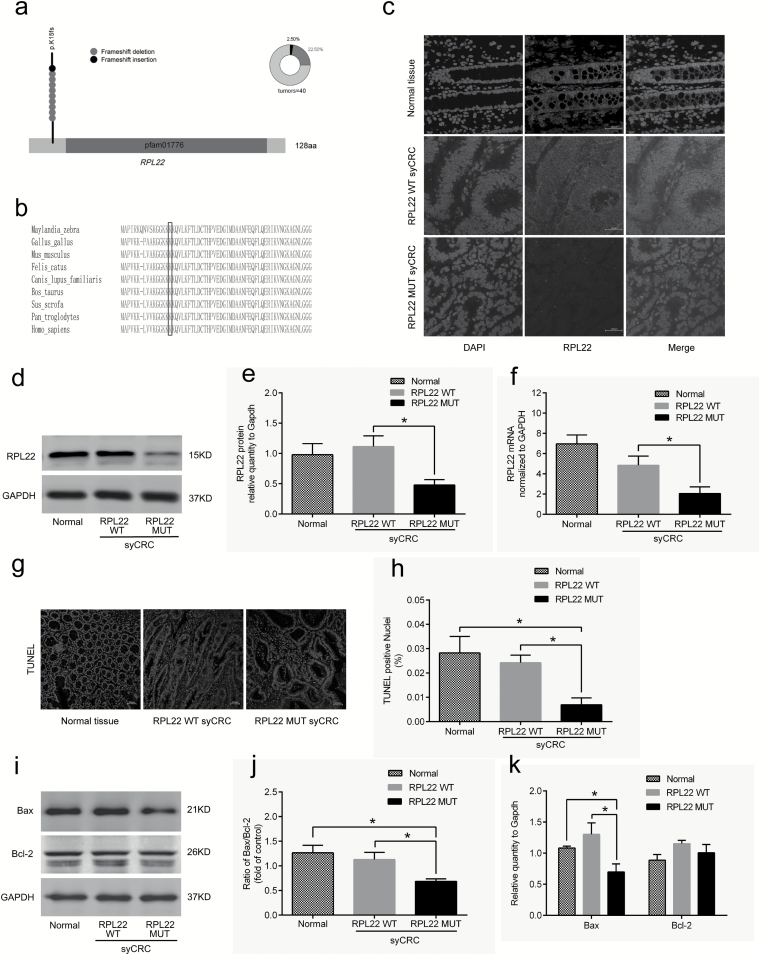
Figure 4.
Characterization of the RPL22 gene in syCRCs. (a) The RPL22 hotspot (p.K15fs) mutation. (b) Conservation of residue K15 in the coding region among near species. (c) Immunofluorescent staining with the RPL22 antibody (red) in normal adjacent tissues and syCRCs with RPL22 WT and RPL22 mutation (MUT; ×40 magnification). DAPI (blue) was used to locate the nuclei of the cells. Scale bars, 100 μm. (d, e) Immunoblotting was used to analyze expression levels of RPL22 protein in adjacent normal tissue and syCRCs with RPL22 WT and RPL22 MUT. GAPDH was used as loading control. (f) Quantitative reverse transcription (qRT)-PCR analysis of RPL22 WT and MUT mRNA expression in syCRCs. (g, h) Representative images of TUNEL positive cells(green) and DAPI (blue) were captured in normal adjacent tissues and syCRCs with RPL22 WT and RPL22 MUT by a fluorescence microscope(×40 magnification). Scale bars, 100 μm. Percentage of TUNEL positive cells in each sample was calculated based in total number of DAPI positive cells for that sample. (i–k) Western blotting analysis of Bax and Bcl-2 protein expression in normal adjacent tissues and syCRCs with RPL22 WT and RPL22 MUT. GAPDH was used as loading control. All experiments were performed at least three independent experiments; Data are mean ± SEM. The P values (determined by the unpaired t-test, two-tailed) relative to normal adjacent tissues or syCRCs with RPL22 WT are shown. *P < 0.05, **P < 0.01, ***P < 0.001. WT, wild-type; MUT, mutation.